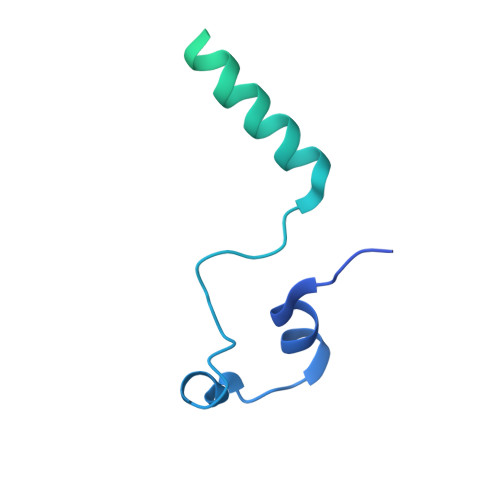
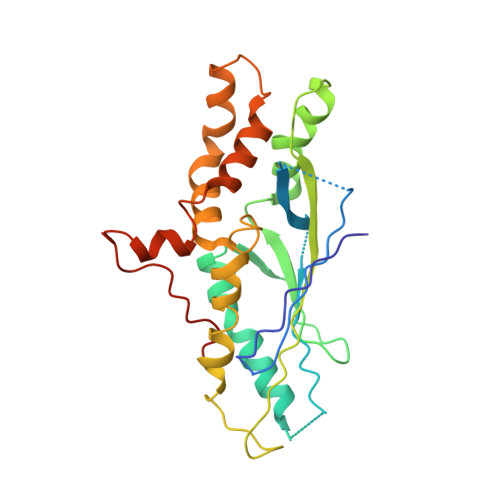
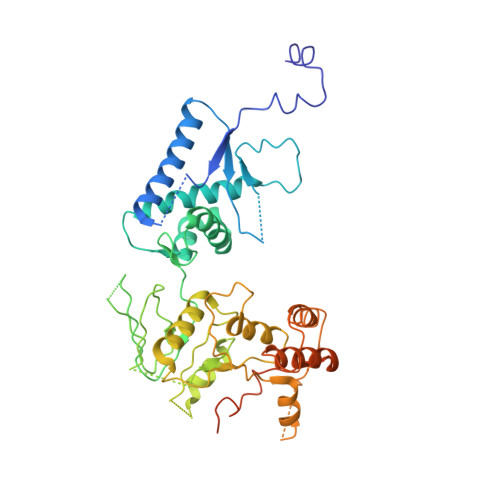
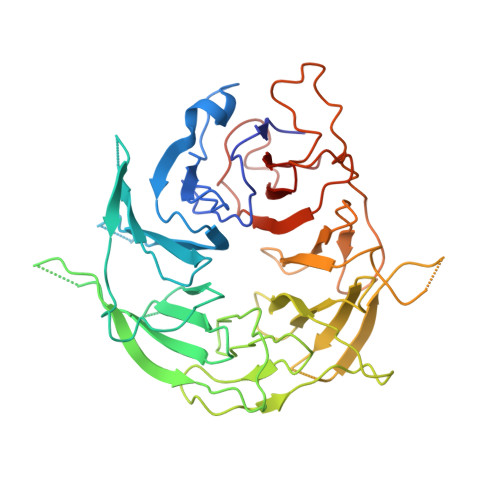
Cryo-EM structures of the Spo11 core complex bound to DNA.
Yu, Y., Wang, J., Liu, K., Zheng, Z., Arter, M., Claeys Bouuaert, C., Pu, S., Patel, D.J., Keeney, S.(2025) Nat Struct Mol Biol 32: 113-124
- PubMed: 39304764
- DOI: https://doi.org/10.1038/s41594-024-01382-8
- Primary Citation of Related Structures:
8URQ, 8URU - PubMed Abstract:
DNA double-strand breaks that initiate meiotic recombination are formed by the topoisomerase-relative enzyme Spo11, supported by conserved auxiliary factors. Because high-resolution structural data have not been available, many questions remain about the architecture of Spo11 and its partners and how they engage with DNA. We report cryo-electron microscopy structures at up to 3.3-Å resolution of DNA-bound core complexes of Saccharomyces cerevisiae Spo11 with Rec102, Rec104 and Ski8. In these structures, monomeric core complexes make extensive contacts with the DNA backbone and with the recessed 3'-OH and first 5' overhanging nucleotide, establishing the molecular determinants of DNA end-binding specificity and providing insight into DNA cleavage preferences in vivo. The structures of individual subunits and their interfaces, supported by functional data in yeast, provide insight into the role of metal ions in DNA binding and uncover unexpected structural variation in homologs of the Top6BL component of the core complex.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Organizational Affiliation: