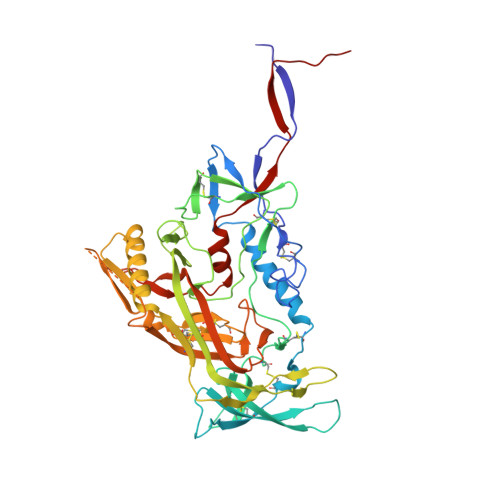
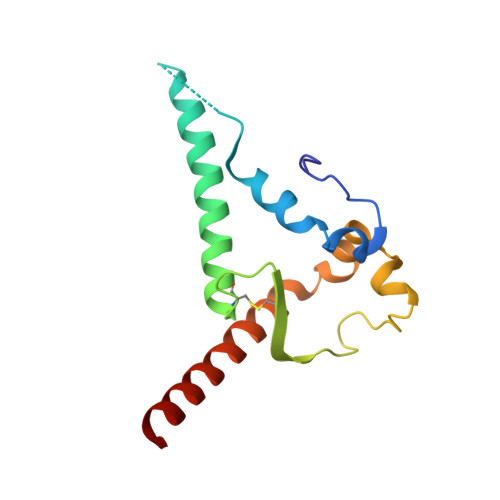
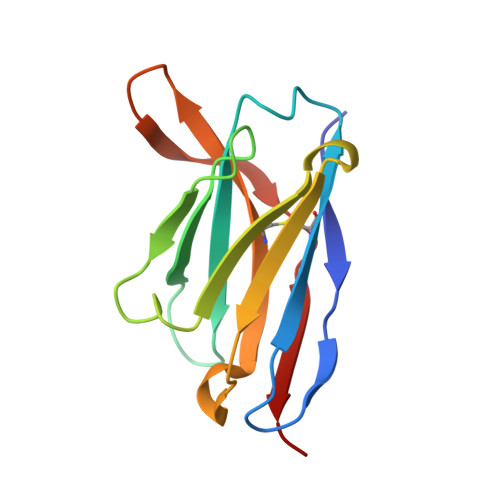
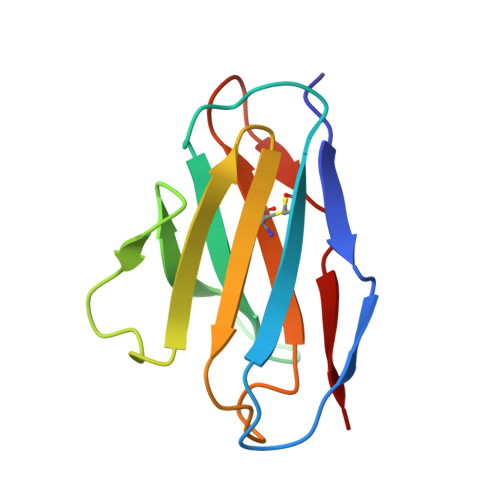
Microsecond dynamics control the HIV-1 Envelope conformation.
Bennett, A.L., Edwards, R., Kosheleva, I., Saunders, C., Bililign, Y., Williams, A., Bubphamala, P., Manosouri, K., Anasti, K., Saunders, K.O., Alam, S.M., Haynes, B.F., Acharya, P., Henderson, R.(2024) Sci Adv 10: eadj0396-eadj0396
- PubMed: 38306419
- DOI: https://doi.org/10.1126/sciadv.adj0396
- Primary Citation of Related Structures:
8SXI, 8SXJ - PubMed Abstract:
The HIV-1 Envelope (Env) glycoprotein facilitates host cell fusion through a complex series of receptor-induced structural changes. Although remarkable progress has been made in understanding the structures of various Env conformations, microsecond timescale dynamics have not been studied experimentally. Here, we used time-resolved, temperature-jump small-angle x-ray scattering to monitor structural rearrangements in an HIV-1 Env SOSIP ectodomain construct with microsecond precision. In two distinct Env variants, we detected a transition that correlated with known Env structure rearrangements with a time constant in the hundreds of microseconds range. A previously unknown structural transition was also observed, which occurred with a time constant below 10 μs, and involved an order-to-disorder transition in the trimer apex. Using this information, we engineered an Env SOSIP construct that locks the trimer in the prefusion closed state by connecting adjacent protomers via disulfides. Our findings show that the microsecond timescale structural dynamics play an essential role in controlling the Env conformation with impacts on vaccine design.
- Duke Human Vaccine Institute, Duke University Medical Center, Durham, NC 27710, USA.
Organizational Affiliation: