Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
Bonazzi, S., Gray, A., Thomsen, N.M., Biag, J., Labbe-Giguere, N., Keaney, E.P., Malik, H.A., Sun, Y., Nunez, J., Karki, R.G., Knapp, M., Elling, R., Fuller, J., Pardee, G., Craig, L., Capre, K., Salas, S., Gorde, A., Liang, G., Lubicka, D., McTighe, S.M., Goold, C., Liu, S., Deng, L., Hong, J., Fekete, A., Stadelmann, P., Frieauff, W., Elhajouji, A., Bauer, D., Lerchner, A., Radetich, B., Furet, P., Piizzi, G., Burdette, D., Wilson, C.J., Peukert, S., Hamann, L.G., Murphy, L.O., Curtis, D.(2023) J Med Chem 66: 9095-9119
- PubMed: 37399505
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00705
- Primary Citation of Related Structures:
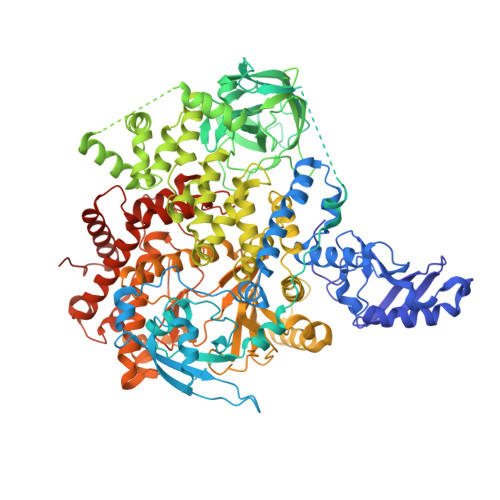
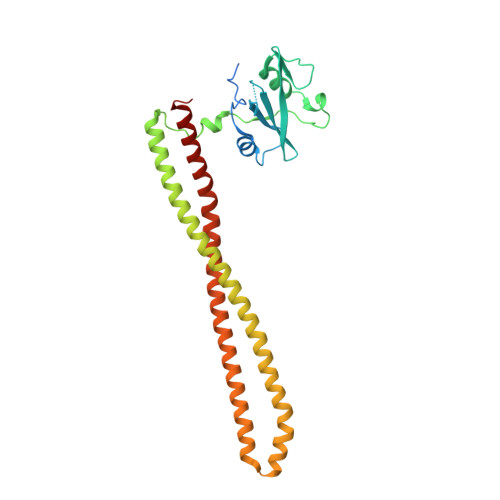
8SBC, 8SBJ - PubMed Abstract:
The allosteric inhibitor of the mechanistic target of rapamycin (mTOR) everolimus reduces seizures in tuberous sclerosis complex (TSC) patients through partial inhibition of mTOR functions. Due to its limited brain permeability, we sought to develop a catalytic mTOR inhibitor optimized for central nervous system (CNS) indications. We recently reported an mTOR inhibitor ( 1 ) that is able to block mTOR functions in the mouse brain and extend the survival of mice with neuronal-specific ablation of the Tsc 1 gene. However, 1 showed the risk of genotoxicity in vitro . Through structure-activity relationship (SAR) optimization, we identified compounds 9 and 11 without genotoxicity risk. In neuronal cell-based models of mTOR hyperactivity, both corrected aberrant mTOR activity and significantly improved the survival rate of mice in the Tsc 1 gene knockout model. Unfortunately, 9 and 11 showed limited oral exposures in higher species and dose-limiting toxicities in cynomolgus macaque, respectively. However, they remain optimal tools to explore mTOR hyperactivity in CNS disease models.
Organizational Affiliation:
Global Discovery Chemistry, Novartis Institutes for BioMedical Research, 181 Massachusetts Ave, Cambridge, Massachusetts 02139, United States.