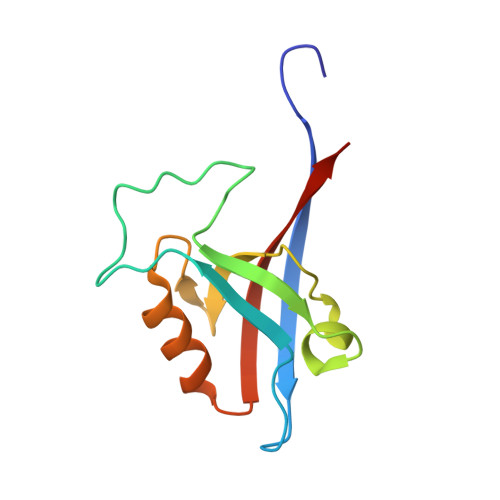
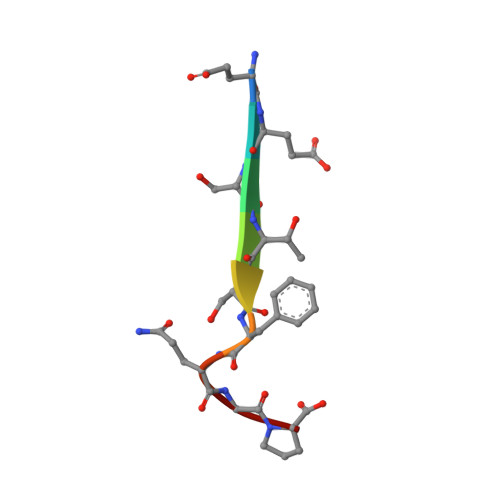
Biophysical and structural analyses of the interaction between the SHANK1 PDZ domain and an internal SLiM.
Li, Y., Trinh, C.H., Acevedo-Jake, A., Gimenez, D., Warriner, S.L., Wilson, A.J.(2024) Biochem J 481: 945-955
- PubMed: 38899489
- DOI: https://doi.org/10.1042/BCJ20240126
- Primary Citation of Related Structures:
8S1R - PubMed Abstract:
The PDZ (Postsynaptic density protein-95[PSD-95]/Discs-large) domain, prevalent as a recognition module, has attracted significant attention given its ability to specifically recognize ligands with consensus motifs (also termed PDZ binding motifs [PBMs]). PBMs typically bear a C-terminal carboxylate as a recognition handle and have been extensively characterised, whilst internal ligands are less well known. Here we characterize a short linear motif (SLiM) - EESTSFQGP - as an internal PBM based on its strong binding affinity towards the SHANK1 PDZ domain (SHANK1656-762 hereafter referred to as SHANK1). Using the acetylated analogue Ac-EESTSFQGP-CONH2 as a competitor for the interaction of SHANK1 with FAM-Ahx-EESTSFQGP-CONH2 or a typical fluorophore-labelled C-terminal PBM - GKAP - FITC-Ahx-EAQTRL-COOH - the internal SLiM was demonstrated to show comparable low-micromolar IC50 by competition fluorescent anisotropy (FA). To gain further insight on the internal ligand interaction at the molecular level, we obtained the X-ray co-crystal structure of the Ac-EESTSFQGP-CONH2/SHANK1 complex and compared this to the Ac-EAQTRL-COOH/SHANK1 complex. The crystallographic studies reveal that the SHANK1 backbones for the two interactions overlap significantly. The main structural differences were shown to result from the flexible loops which reorganise to accommodate the two PBMs with distinct lengths and terminal groups. In addition, the two C-terminal residues Gly and Pro in Ac-EESTSFQGP-CONH2 were shown not to participate in interaction with the target protein, implying further truncation and structural modification using peptidomimetic approaches on this sequence may be feasible. Taken together, the SLiM Ac-EESTSFQGP-CONH2 holds potential as an internal ligand for targeting SHANK1.
- University of Birmingham, Birmingham, United Kingdom.
Organizational Affiliation: