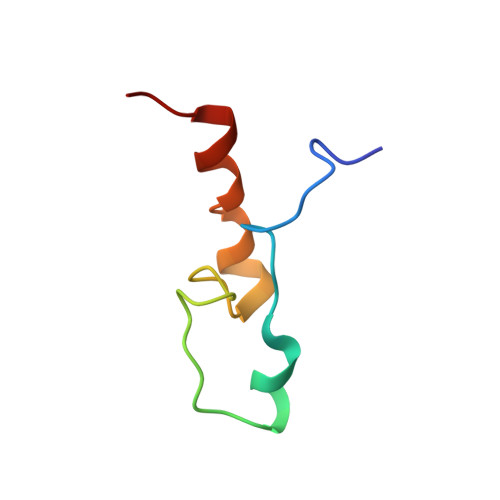
Structure and RNA-binding of the helically extended Roquin CCCH-type zinc finger.
Tants, J.N., Oberstrass, L., Weigand, J.E., Schlundt, A.(2024) Nucleic Acids Res 52: 9838-9853
- PubMed: 38953172
- DOI: https://doi.org/10.1093/nar/gkae555
- Primary Citation of Related Structures:
8RHS - PubMed Abstract:
Zinc finger (ZnF) domains appear in a pool of structural contexts and despite their small size achieve varying target specificities, covering single-stranded and double-stranded DNA and RNA as well as proteins. Combined with other RNA-binding domains, ZnFs enhance affinity and specificity of RNA-binding proteins (RBPs). The ZnF-containing immunoregulatory RBP Roquin initiates mRNA decay, thereby controlling the adaptive immune system. Its unique ROQ domain shape-specifically recognizes stem-looped cis-elements in mRNA 3'-untranslated regions (UTR). The N-terminus of Roquin contains a RING domain for protein-protein interactions and a ZnF, which was suggested to play an essential role in RNA decay by Roquin. The ZnF domain boundaries, its RNA motif preference and its interplay with the ROQ domain have remained elusive, also driven by the lack of high-resolution data of the challenging protein. We provide the solution structure of the Roquin-1 ZnF and use an RBNS-NMR pipeline to show that the ZnF recognizes AU-rich RNAs. We systematically refine the contributions of adenines in a poly(U)-background to specific complex formation. With the simultaneous binding of ROQ and ZnF to a natural target transcript of Roquin, our study for the first time suggests how Roquin integrates RNA shape and sequence features through the ROQ-ZnF tandem.
- Institute for Molecular Biosciences and Biomolecular Resonance Center (BMRZ), Goethe University Frankfurt, Max-von-Laue-Str. 7-9, 60438 Frankfurt, Germany.
Organizational Affiliation: