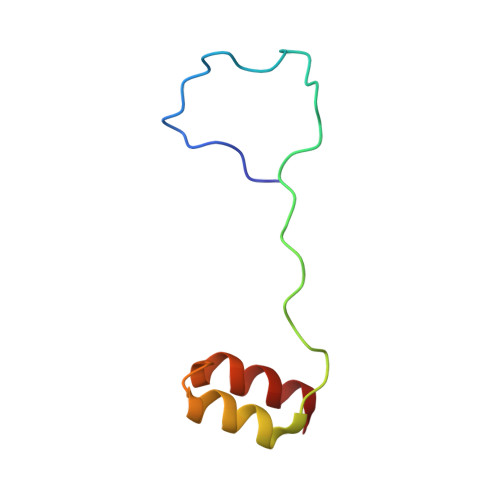
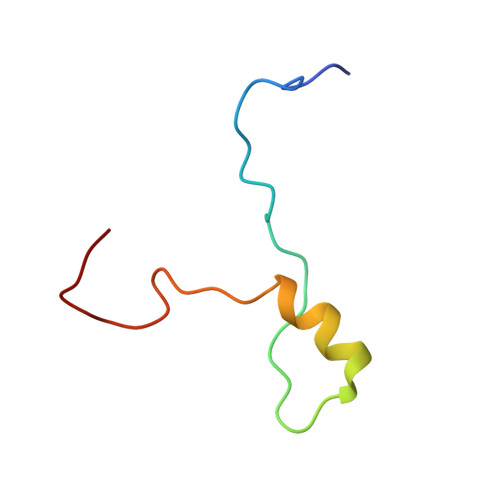
Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Scat, S., Weissman, K.J., Chagot, B.(2024) RSC Chem Biol 5: 669-683
- PubMed: 38966669
- DOI: https://doi.org/10.1039/d4cb00075g
- Primary Citation of Related Structures:
8RAJ - PubMed Abstract:
The fidelity of biosynthesis by modular polyketide synthases (PKSs) depends on specific moderate affinity interactions between successive polypeptide subunits mediated by docking domains (DDs). These sequence elements are notably portable, allowing their transplantation into alternative biosynthetic and metabolic contexts. Herein, we use integrative structural biology to characterize a pair of DDs from the toblerol trans -AT PKS. Both are intrinsically disordered regions (IDRs) that fold into a 3 α-helix docking complex of unprecedented topology. The C-terminal docking domain ( C DD) resembles the 4 α-helix type (4HB) C DDs, which shows that the same type of DD can be redeployed to form complexes of distinct geometry. By carefully re-examining known DD structures, we further extend this observation to type 2 docking domains, establishing previously unsuspected structural relations between DD types. Taken together, these data illustrate the plasticity of α-helical DDs, which allow the formation of a diverse topological spectrum of docked complexes. The newly identified DDs should also find utility in modular PKS genetic engineering.
- Université de Lorraine, CNRS, IMoPA F-54000 Nancy France kira.weissman@univ-lorraine.fr benjamin.chagot@univ-lorraine.fr.
Organizational Affiliation: