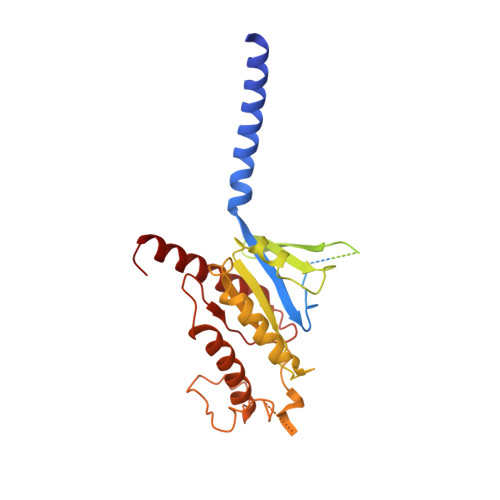
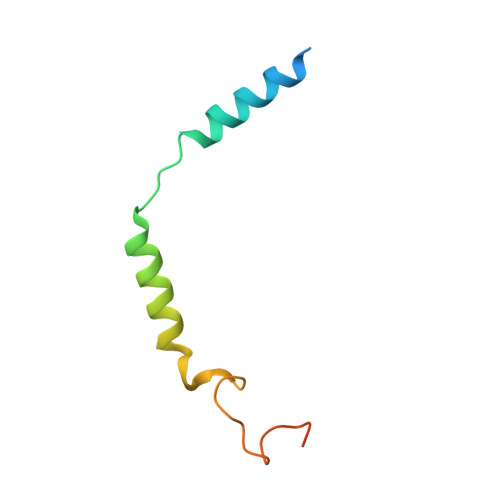
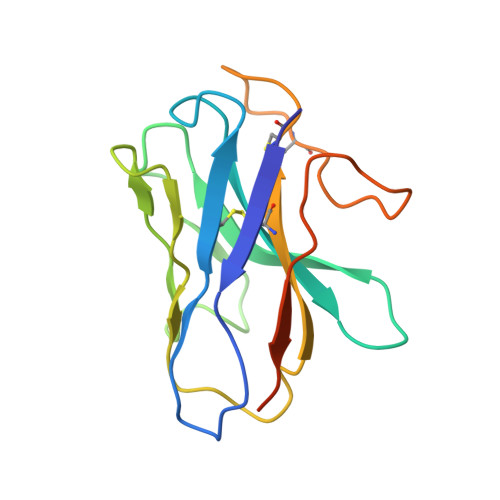
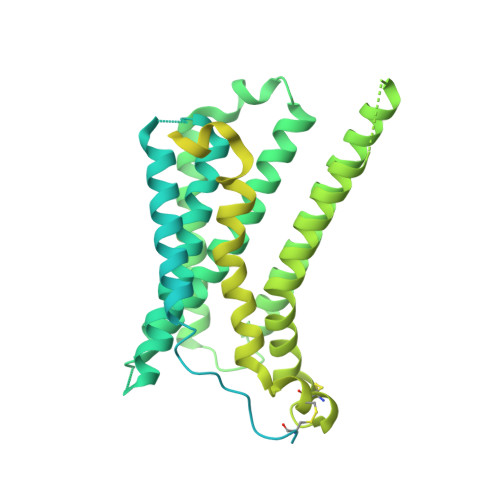
Structure elucidation of a human melanocortin-4 receptor specific orthosteric nanobody agonist.
Fontaine, T., Busch, A., Laeremans, T., De Cesco, S., Liang, Y.L., Jaakola, V.P., Sands, Z., Triest, S., Masiulis, S., Dekeyzer, L., Samyn, N., Loeys, N., Perneel, L., Debaere, M., Martini, M., Vantieghem, C., Virmani, R., Skieterska, K., Staelens, S., Barroco, R., Van Roy, M., Menet, C.(2024) Nat Commun 15: 7029-7029
- PubMed: 39353917
- DOI: https://doi.org/10.1038/s41467-024-50827-7
- Primary Citation of Related Structures:
8QJ2 - PubMed Abstract:
The melanocortin receptor 4 (MC4R) belongs to the melanocortin receptor family of G-protein coupled receptors and is a key switch in the leptin-melanocortin molecular axis that controls hunger and satiety. Brain-produced hormones such as α-melanocyte-stimulating hormone (agonist) and agouti-related peptide (inverse agonist) regulate the molecular communication of the MC4R axis but are promiscuous for melanocortin receptor subtypes and induce a wide array of biological effects. Here, we use a chimeric construct of conformation-selective, nanobody-based binding domain (a ConfoBody Cb80) and active state-stabilized MC4R-β2AR hybrid for efficient de novo discovery of a sequence diverse panel of MC4R-specific, potent and full agonistic nanobodies. We solve the active state MC4R structure in complex with the full agonistic nanobody pN162 at 3.4 Å resolution. The structure shows a distinct interaction with pN162 binding deeply in the orthosteric pocket. MC4R peptide agonists, such as the marketed setmelanotide, lack receptor selectivity and show off-target effects. In contrast, the agonistic nanobody is highly specific and hence can be a more suitable agent for anti-obesity therapeutic intervention via MC4R.
- Confo Therapeutics N.V, Ghent, Belgium.
Organizational Affiliation: