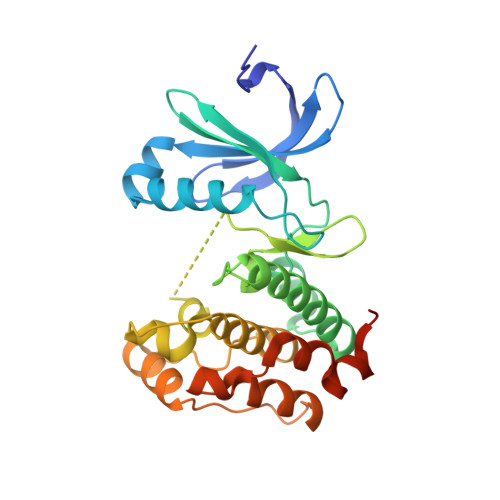
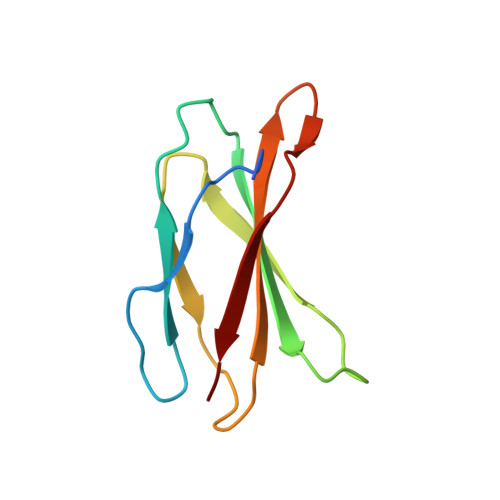
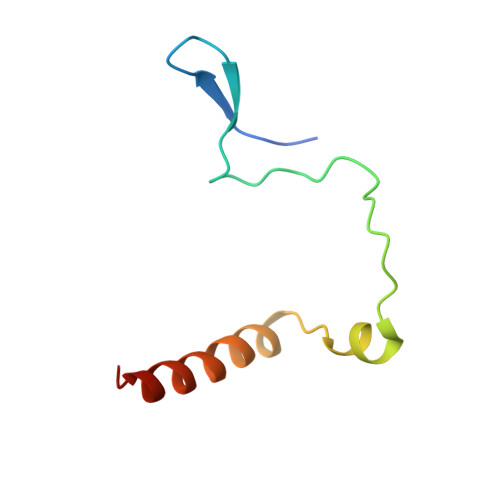
CEP192 localises mitotic Aurora-A activity by priming its interaction with TPX2.
Holder, J., Miles, J.A., Batchelor, M., Popple, H., Walko, M., Yeung, W., Kannan, N., Wilson, A.J., Bayliss, R., Gergely, F.(2024) EMBO J 43: 5381-5420
- PubMed: 39327527
- DOI: https://doi.org/10.1038/s44318-024-00240-z
- Primary Citation of Related Structures:
8PR7 - PubMed Abstract:
Aurora-A is an essential cell-cycle kinase with critical roles in mitotic entry and spindle dynamics. These functions require binding partners such as CEP192 and TPX2, which modulate both kinase activity and localisation of Aurora-A. Here we investigate the structure and role of the centrosomal Aurora-A:CEP192 complex in the wider molecular network. We find that CEP192 wraps around Aurora-A, occupies the binding sites for mitotic spindle-associated partners, and thus competes with them. Comparison of two different Aurora-A conformations reveals how CEP192 modifies kinase activity through the site used for TPX2-mediated activation. Deleting the Aurora-A-binding interface in CEP192 prevents centrosomal accumulation of Aurora-A, curtails its activation-loop phosphorylation, and reduces spindle-bound TPX2:Aurora-A complexes, resulting in error-prone mitosis. Thus, by supplying the pool of phosphorylated Aurora-A necessary for TPX2 binding, CEP192:Aurora-A complexes regulate spindle function. We propose an evolutionarily conserved spatial hierarchy, which protects genome integrity through fine-tuning and correctly localising Aurora-A activity.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: