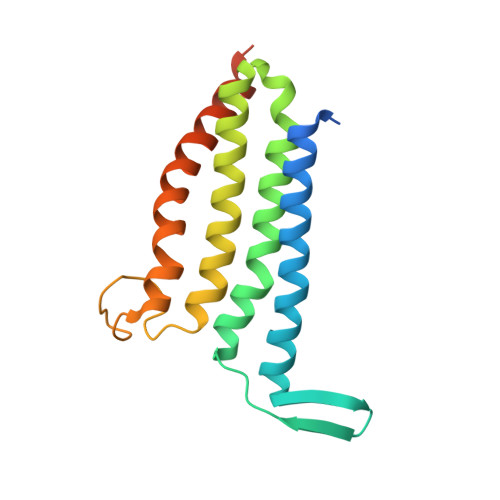
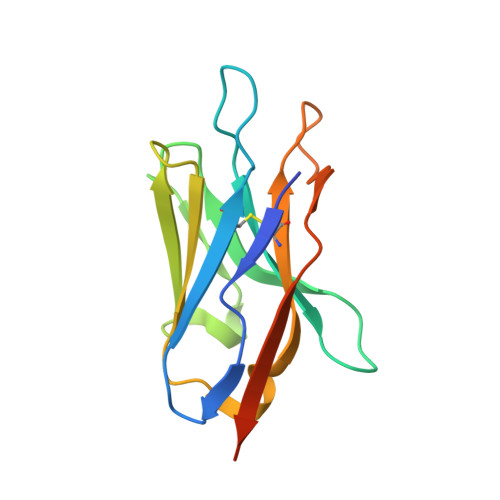
Structural basis of the Meinwald rearrangement catalysed by styrene oxide isomerase.
Khanppnavar, B., Choo, J.P.S., Hagedoorn, P.L., Smolentsev, G., Stefanic, S., Kumaran, S., Tischler, D., Winkler, F.K., Korkhov, V.M., Li, Z., Kammerer, R.A., Li, X.(2024) Nat Chem 16: 1496-1504
- PubMed: 38744914
- DOI: https://doi.org/10.1038/s41557-024-01523-y
- Primary Citation of Related Structures:
8PNU, 8PNV - PubMed Abstract:
Membrane-bound styrene oxide isomerase (SOI) catalyses the Meinwald rearrangement-a Lewis-acid-catalysed isomerization of an epoxide to a carbonyl compound-and has been used in single and cascade reactions. However, the structural information that explains its reaction mechanism has remained elusive. Here we determine cryo-electron microscopy (cryo-EM) structures of SOI bound to a single-domain antibody with and without the competitive inhibitor benzylamine, and elucidate the catalytic mechanism using electron paramagnetic resonance spectroscopy, functional assays, biophysical methods and docking experiments. We find ferric haem b bound at the subunit interface of the trimeric enzyme through H58, where Fe(III) acts as the Lewis acid by binding to the epoxide oxygen. Y103 and N64 and a hydrophobic pocket binding the oxygen of the epoxide and the aryl group, respectively, position substrates in a manner that explains the high regio-selectivity and stereo-specificity of SOI. Our findings can support extending the range of epoxide substrates and be used to potentially repurpose SOI for the catalysis of new-to-nature Fe-based chemical reactions.
- Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institute, Villigen, Switzerland.
Organizational Affiliation: