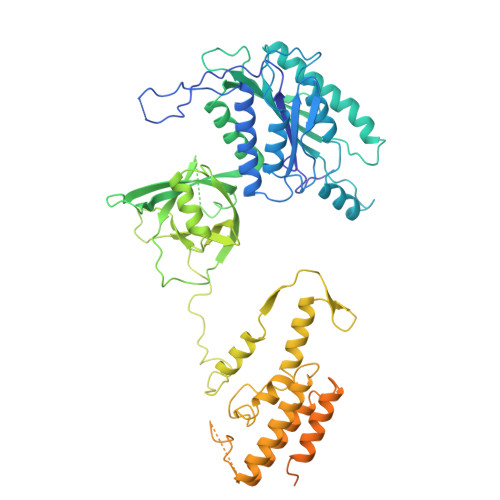
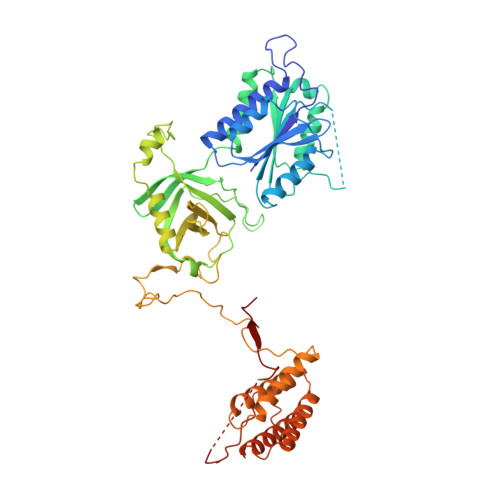
Basis of gene-specific transcription regulation by the Integrator complex.
Sabath, K., Nabih, A., Arnold, C., Moussa, R., Domjan, D., Zaugg, J.B., Jonas, S.(2024) Mol Cell 84: 2525
- PubMed: 38906142
- DOI: https://doi.org/10.1016/j.molcel.2024.05.027
- Primary Citation of Related Structures:
8PK5, 8PK6 - PubMed Abstract:
The Integrator complex attenuates gene expression via the premature termination of RNA polymerase II (RNAP2) at promoter-proximal pausing sites. It is required for stimulus response, cell differentiation, and neurodevelopment, but how gene-specific and adaptive regulation by Integrator is achieved remains unclear. Here, we identify two sites on human Integrator subunits 13/14 that serve as binding hubs for sequence-specific transcription factors (TFs) and other transcription effector complexes. When Integrator is attached to paused RNAP2, these hubs are positioned upstream of the transcription bubble, consistent with simultaneous TF-promoter tethering. The TFs co-localize with Integrator genome-wide, increase Integrator abundance on target genes, and co-regulate responsive transcriptional programs. For instance, sensory cilia formation induced by glucose starvation depends on Integrator-TF contacts. Our data suggest TF-mediated promoter recruitment of Integrator as a widespread mechanism for targeted transcription regulation.
- Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, 8093 Zurich, Switzerland. Electronic address: kevin.sabath@imp.ac.at.
Organizational Affiliation: