Structure basis of translation regulation by YchF bound to ribosome
Li, X., Yu, T., Li, Q., Zeng, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosome-binding ATPase YchF | A [auth y] | 379 | Escherichia coli | Mutation(s): 1 Gene Names: ychF | 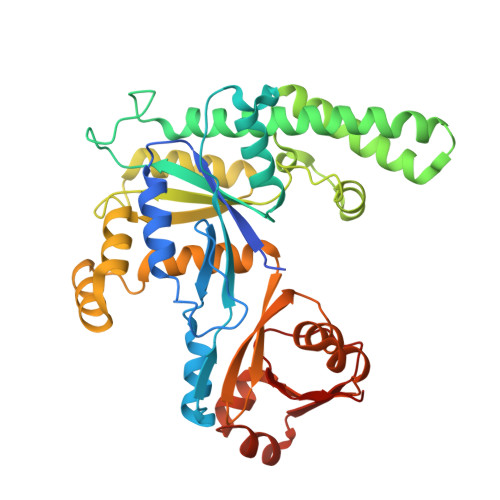 |
UniProt | |||||
Find proteins for P0ABU2 (Escherichia coli (strain K12)) Explore P0ABU2 Go to UniProtKB: P0ABU2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ABU2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L14 | C [auth j] | 123 | Escherichia coli | Mutation(s): 0 | 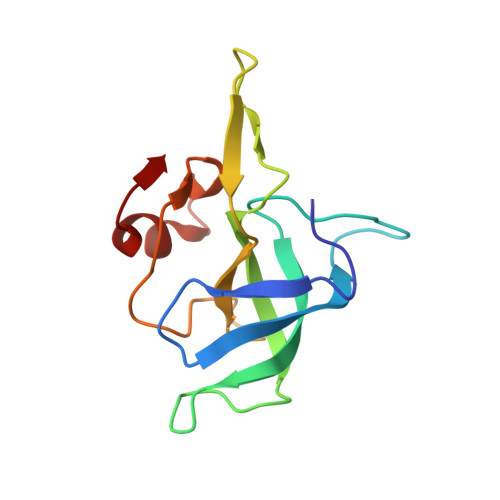 |
UniProt | |||||
Find proteins for P0ADY3 (Escherichia coli (strain K12)) Explore P0ADY3 Go to UniProtKB: P0ADY3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADY3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L19 | D [auth o] | 115 | Escherichia coli | Mutation(s): 0 | 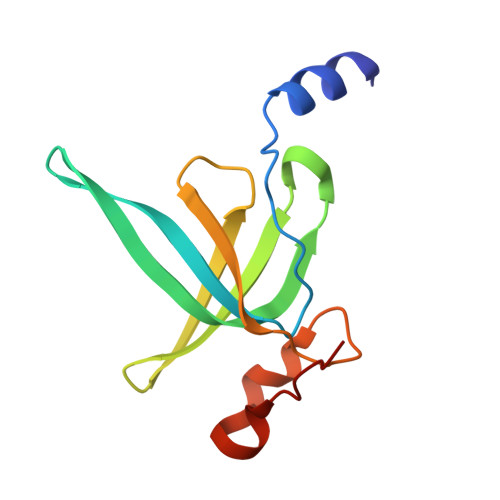 |
UniProt | |||||
Find proteins for P0A7K6 (Escherichia coli (strain K12)) Explore P0A7K6 Go to UniProtKB: P0A7K6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7K6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 23S rRNA (partial) | B [auth a] | 2,904 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | AA [auth a] BA [auth a] CA [auth a] DA [auth a] E [auth a] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32171200 |
| Guangdong Basic and Applied Basic Research Foundation | China | 2021A1515010805 |
| Shenzhen Science and Technology Program | China | JCYJ20220530115210023 |