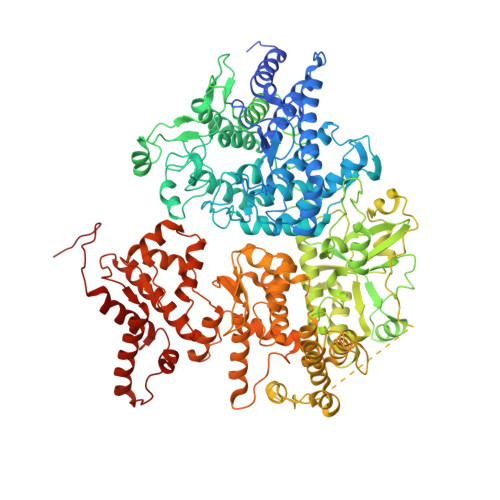
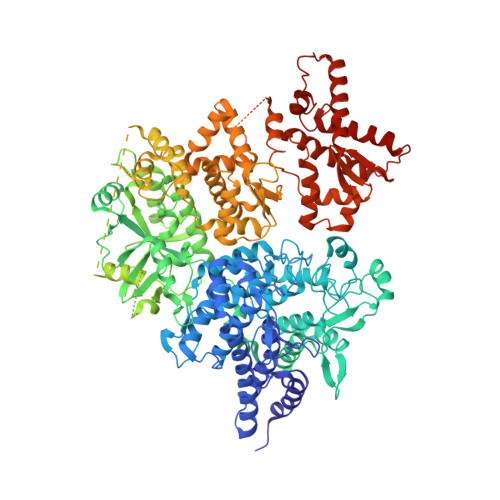
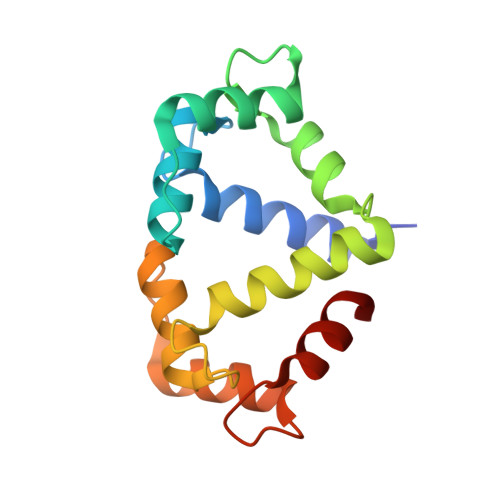
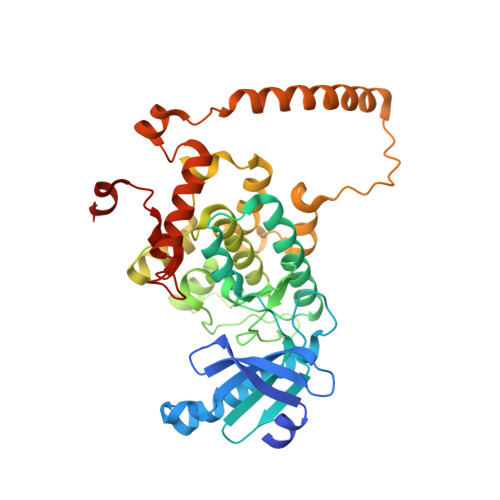
Architecture and activation of human muscle phosphorylase kinase.
Yang, X., Zhu, M., Lu, X., Wang, Y., Xiao, J.(2024) Nat Commun 15: 2719-2719
- PubMed: 38548794
- DOI: https://doi.org/10.1038/s41467-024-47049-2
- Primary Citation of Related Structures:
8JFK, 8JFL, 8XY7, 8XYA, 8XYB - PubMed Abstract:
The study of phosphorylase kinase (PhK)-regulated glycogen metabolism has contributed to the fundamental understanding of protein phosphorylation; however, the molecular mechanism of PhK remains poorly understood. Here we present the high-resolution cryo-electron microscopy structures of human muscle PhK. The 1.3-megadalton PhK α 4 β 4 γ 4 δ 4 hexadecamer consists of a tetramer of tetramer, wherein four αβγδ modules are connected by the central β 4 scaffold. The α- and β-subunits possess glucoamylase-like domains, but exhibit no detectable enzyme activities. The α-subunit serves as a bridge between the β-subunit and the γδ subcomplex, and facilitates the γ-subunit to adopt an autoinhibited state. Ca 2+ -free calmodulin (δ-subunit) binds to the γ-subunit in a compact conformation. Upon binding of Ca 2+ , a conformational change occurs, allowing for the de-inhibition of the γ-subunit through a spring-loaded mechanism. We also reveal an ADP-binding pocket in the β-subunit, which plays a role in allosterically enhancing PhK activity. These results provide molecular insights of this important kinase complex.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, P.R. China.
Organizational Affiliation: