Crystal structure of pilin-specific sortase SrtC2 from Lactobacillus rhamnosus GG
Das, S., Megta, A.K., Pratap, S., Krishnan, V.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Class C sortase | 152 | Lacticaseibacillus rhamnosus GG | Mutation(s): 0 Gene Names: GM657_11825, LRHM_2278 | 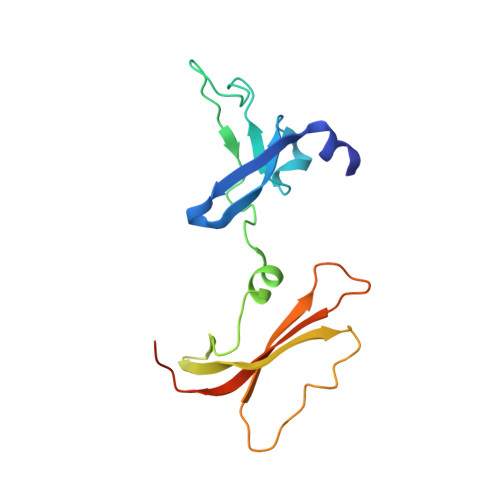 | |
UniProt | |||||
Find proteins for A0A7S7JH80 (Lacticaseibacillus rhamnosus (strain ATCC 53103 / LMG 18243 / GG)) Explore A0A7S7JH80 Go to UniProtKB: A0A7S7JH80 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7S7JH80 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 80.4 | α = 90 |
| b = 80.4 | β = 90 |
| c = 46.895 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| STARANISO | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Biotechnology (DBT, India) | India | BT/PR5891/BRB/10/1098/2012 |