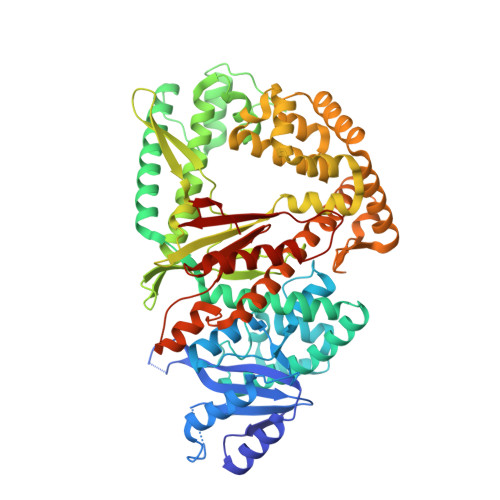
Discovery of a small-molecule inhibitor that traps Pol theta on DNA and synergizes with PARP inhibitors.
Fried, W., Tyagi, M., Minakhin, L., Chandramouly, G., Tredinnick, T., Ramanjulu, M., Auerbacher, W., Calbert, M., Rusanov, T., Hoang, T., Borisonnik, N., Betsch, R., Krais, J.J., Wang, Y., Vekariya, U.M., Gordon, J., Morton, G., Kent, T., Skorski, T., Johnson, N., Childers, W., Chen, X.S., Pomerantz, R.T.(2024) Nat Commun 15: 2862-2862
- PubMed: 38580648
- DOI: https://doi.org/10.1038/s41467-024-46593-1
- Primary Citation of Related Structures:
8GD7 - PubMed Abstract:
The DNA damage response (DDR) protein DNA Polymerase θ (Polθ) is synthetic lethal with homologous recombination (HR) factors and is therefore a promising drug target in BRCA1/2 mutant cancers. We discover an allosteric Polθ inhibitor (Polθi) class with 4-6 nM IC 50 that selectively kills HR-deficient cells and acts synergistically with PARP inhibitors (PARPi) in multiple genetic backgrounds. X-ray crystallography and biochemistry reveal that Polθi selectively inhibits Polθ polymerase (Polθ-pol) in the closed conformation on B-form DNA/DNA via an induced fit mechanism. In contrast, Polθi fails to inhibit Polθ-pol catalytic activity on A-form DNA/RNA in which the enzyme binds in the open configuration. Remarkably, Polθi binding to the Polθ-pol:DNA/DNA closed complex traps the polymerase on DNA for more than forty minutes which elucidates the inhibitory mechanism of action. These data reveal a unique small-molecule DNA polymerase:DNA trapping mechanism that induces synthetic lethality in HR-deficient cells and potentiates the activity of PARPi.
Organizational Affiliation:
Molecular and Computational Biology, Department of Biological Sciences and Chemistry, University of Southern California, Los Angeles, CA, USA.