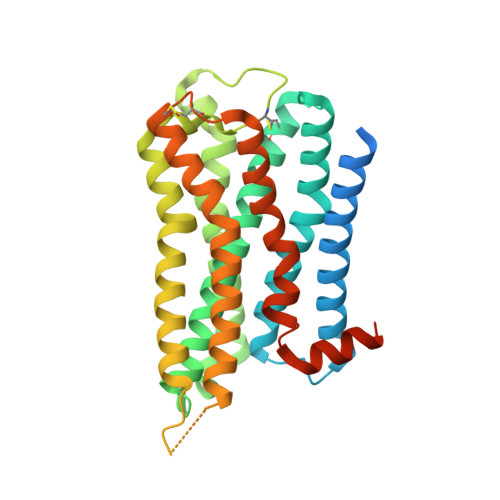
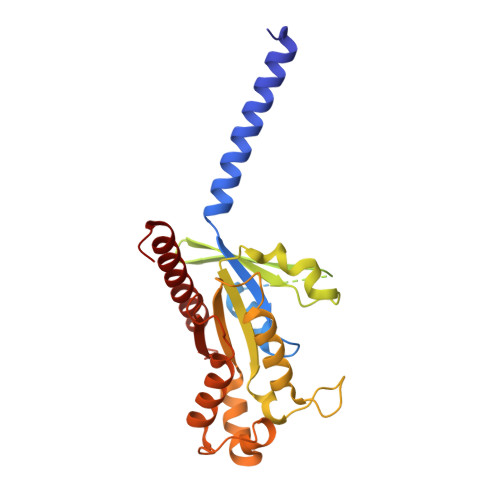
Xanomeline displays concomitant orthosteric and allosteric binding modes at the M 4 mAChR.
Burger, W.A.C., Pham, V., Vuckovic, Z., Powers, A.S., Mobbs, J.I., Laloudakis, Y., Glukhova, A., Wootten, D., Tobin, A.B., Sexton, P.M., Paul, S.M., Felder, C.C., Danev, R., Dror, R.O., Christopoulos, A., Valant, C., Thal, D.M.(2023) Nat Commun 14: 5440-5440
- PubMed: 37673901
- DOI: https://doi.org/10.1038/s41467-023-41199-5
- Primary Citation of Related Structures:
8FX5 - PubMed Abstract:
The M 4 muscarinic acetylcholine receptor (M 4 mAChR) has emerged as a drug target of high therapeutic interest due to its expression in regions of the brain involved in the regulation of psychosis, cognition, and addiction. The mAChR agonist, xanomeline, has provided significant improvement in the Positive and Negative Symptom Scale (PANSS) scores in a Phase II clinical trial for the treatment of patients suffering from schizophrenia. Here we report the active state cryo-EM structure of xanomeline bound to the human M 4 mAChR in complex with the heterotrimeric G i1 transducer protein. Unexpectedly, two molecules of xanomeline were found to concomitantly bind to the monomeric M 4 mAChR, with one molecule bound in the orthosteric (acetylcholine-binding) site and a second molecule in an extracellular vestibular allosteric site. Molecular dynamic simulations supports the structural findings, and pharmacological validation confirmed that xanomeline acts as a dual orthosteric and allosteric ligand at the human M 4 mAChR. These findings provide a basis for further understanding xanomeline's complex pharmacology and highlight the myriad of ways through which clinically relevant ligands can bind to and regulate GPCRs.
- Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC, 3052, Australia.
Organizational Affiliation: