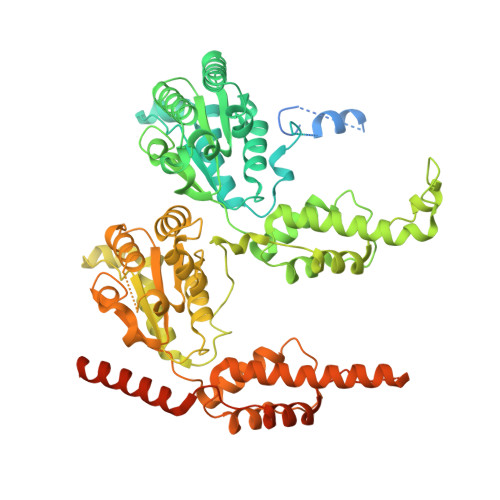
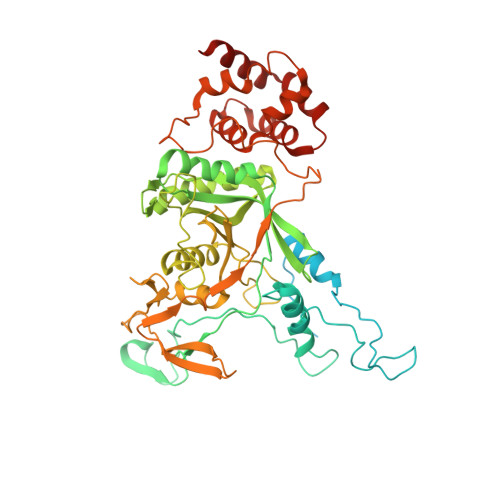
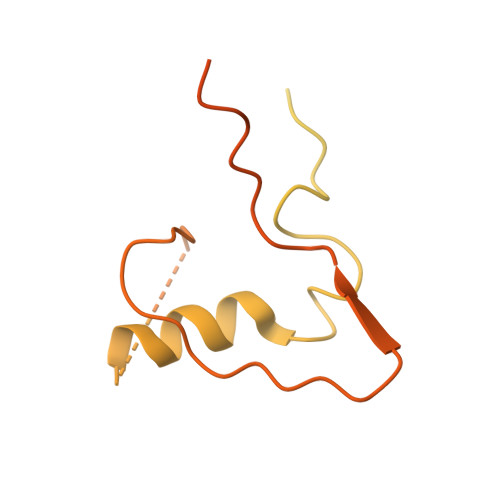
SUMO enhances unfolding of SUMO-polyubiquitin-modified substrates by the Ufd1/Npl4/Cdc48 complex.
Lee, H.G., Lemmon, A.A., Lima, C.D.(2023) Proc Natl Acad Sci U S A 120: e2213703120-e2213703120
- PubMed: 36574706
- DOI: https://doi.org/10.1073/pnas.2213703120
- Primary Citation of Related Structures:
8DAR, 8DAS, 8DAT, 8DAU, 8DAV, 8DAW - PubMed Abstract:
The Ufd1/Npl4/Cdc48 complex is a universal protein segregase that plays key roles in eukaryotic cellular processes. Its functions orchestrating the clearance or removal of polyubiquitylated targets are established; however, prior studies suggest that the complex also targets substrates modified by the ubiquitin-like protein SUMO. Here, we show that interactions between Ufd1 and SUMO enhance unfolding of substrates modified by SUMO-polyubiquitin hybrid chains by the budding yeast Ufd1/Npl4/Cdc48 complex compared to substrates modified by polyubiquitin chains, a difference that is accentuated when the complex has a choice between these substrates. Incubating Ufd1/Npl4/Cdc48 with a substrate modified by a SUMO-polyubiquitin hybrid chain produced a series of single-particle cryo-EM structures that reveal features of interactions between Ufd1/Npl4/Cdc48 and ubiquitin prior to and during unfolding of ubiquitin. These results are consistent with cellular functions for SUMO and ubiquitin modifications and support a physical model wherein Ufd1/Npl4/Cdc48, SUMO, and ubiquitin conjugation pathways converge to promote clearance of proteins modified with SUMO and polyubiquitin.
- Biochemistry, Structural Biology, Cell Biology, Developmental Biology and Molecular Biology (BCMB) Allied Program, Weill Graduate School of Medical Sciences, Cornell University, New York, NY 10065.
Organizational Affiliation: