Structure of anellovirus-like particles reveal a mechanism for immune evasion.
Liou, S.H., Boggavarapu, R., Cohen, N.R., Zhang, Y., Sharma, I., Zeheb, L., Mukund Acharekar, N., Rodgers, H.D., Islam, S., Pitts, J., Arze, C., Swaminathan, H., Yozwiak, N., Ong, T., Hajjar, R.J., Chang, Y., Swanson, K.A., Delagrave, S.(2024) Nat Commun 15: 7219-7219
- PubMed: 39174507
- DOI: https://doi.org/10.1038/s41467-024-51064-8
- Primary Citation of Related Structures:
8CYG, 8V7X - PubMed Abstract:
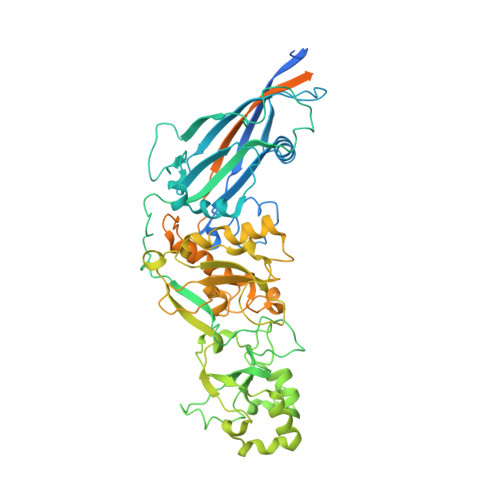
Anelloviruses are nonpathogenic viruses that comprise a major portion of the human virome. Despite being ubiquitous in the human population, anelloviruses (ANVs) remain poorly understood. Basic features of the virus, such as the identity of its capsid protein and the structure of the viral particle, have been unclear until now. Here, we use cryogenic electron microscopy to describe the first structure of an ANV-like particle. The particle, formed by 60 jelly roll domain-containing ANV capsid proteins, forms an icosahedral particle core from which spike domains extend to form a salient part of the particle surface. The spike domains come together around the 5-fold symmetry axis to form crown-like features. The base of the spike domain, the P1 subdomain, shares some sequence conservation between ANV strains while a hypervariable region, forming the P2 subdomain, is at the spike domain apex. We propose that this structure renders the particle less susceptible to antibody neutralization by hiding vulnerable conserved domains while exposing highly diverse epitopes as immunological decoys, thereby contributing to the immune evasion properties of anelloviruses. These results shed light on the structure of anelloviruses and provide a framework to understand their interactions with the immune system.
- Ring Therapeutics, 140 First Street Suite 300, Cambridge, MA, 02139, USA.
Organizational Affiliation: