Cytosine analogues as DNA methyltransferase substrates.
Wojciechowski, M., Czapinska, H., Krwawicz, J., Rafalski, D., Bochtler, M.(2024) Nucleic Acids Res
- PubMed: 38966999
- DOI: https://doi.org/10.1093/nar/gkae568
- Primary Citation of Related Structures:
8C56, 8C57, 8C58, 8C59 - PubMed Abstract:
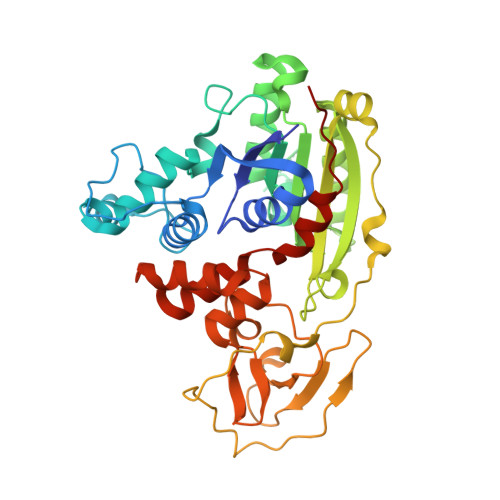
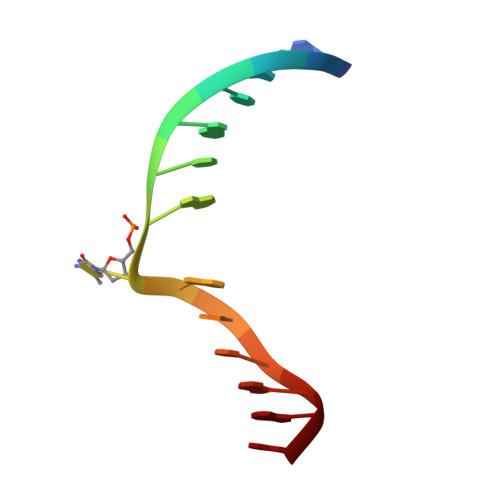
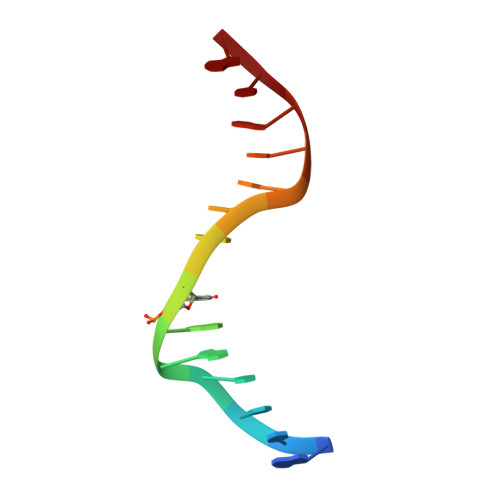
DNA methyltransferases are drug targets for myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), acute myelogenous leukemia (AML) and possibly β-hemoglobinopathies. We characterize the interaction of nucleoside analogues in DNA with a prokaryotic CpG-specific DNA methyltransferase (M.MpeI) as a model for mammalian DNMT1 methyltransferases. We tested DNA containing 5-hydroxymethylcytosine (5hmC), 5-hydroxycytosine (5OHC), 5-methyl-2-pyrimidinone (in the ribosylated form known as 5-methylzebularine, 5mZ), 5,6-dihydro-5-azacytosine (dhaC), 5-fluorocytosine (5FC), 5-chlorocytosine (5ClC), 5-bromocytosine (5BrC) and 5-iodocytosine (5IC). Covalent complex formation was by far most efficient for 5FC. Non-covalent complexes were most abundant for dhaC and 5mZ. Surprisingly, we observed methylation of 5IC and 5BrC, and to a lesser extent 5ClC and 5FC, in the presence, but not the absence of small molecule thiol nucleophiles. For 5IC and 5BrC, we demonstrated by mass spectrometry that the reactions were due to methyltransferase driven dehalogenation, followed by methylation. Crystal structures of M.MpeI-DNA complexes capture the 'in' conformation of the active site loop for analogues with small or rotatable (5mZ) 5-substituents and its 'out' form for bulky 5-substituents. Since very similar 'in' and 'out' loop conformations were also observed for DNMT1, it is likely that our conclusions generalize to other DNA methyltransferases.
Organizational Affiliation:
International Institute of Molecular and Cell Biology, Trojdena 4, 02-109 Warsaw, Poland.