Crystal structure of WT p38alpha
Pous, J., Baginski, B., Gonzalez, L., Macias, M.J., Nebreda, A.R.(null) Res Sq
Experimental Data Snapshot
(null) Res Sq
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitogen-activated protein kinase 14 | A [auth AAA] | 359 | Mus musculus | Mutation(s): 0 Gene Names: Mapk14, Crk1, Csbp1, Csbp2 EC: 2.7.11.24 | 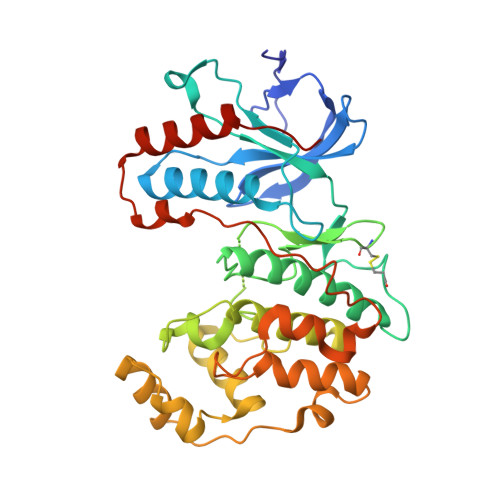 |
UniProt | |||||
Find proteins for P47811 (Mus musculus) Explore P47811 Go to UniProtKB: P47811 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P47811 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SB2 (Subject of Investigation/LOI) Query on SB2 | B [auth AAA] | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE C21 H16 F N3 O S CDMGBJANTYXAIV-MHZLTWQESA-N |  | ||
| MG Query on MG | C [auth AAA], D [auth AAA] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 65.3 | α = 90 |
| b = 74.612 | β = 90 |
| c = 78.1 | γ = 90 |
| Software Name | Purpose |
|---|---|
| iMOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Spanish Ministry of Science, Innovation, and Universities | Spain | PID2019-109521RB-100 |
| La Caixa Foundation | Spain | BioMedTec VIII - Nebre |