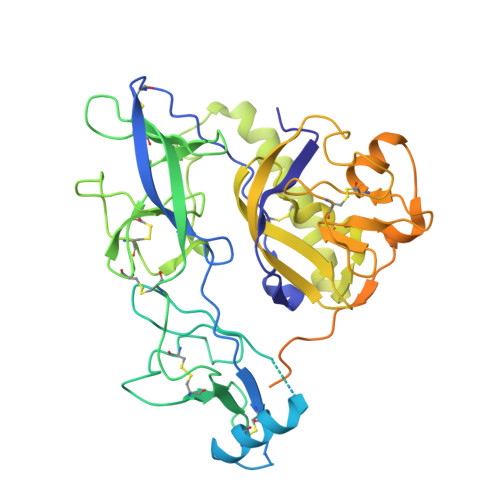
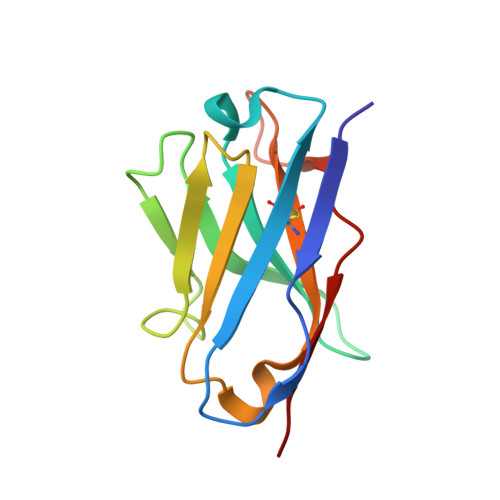
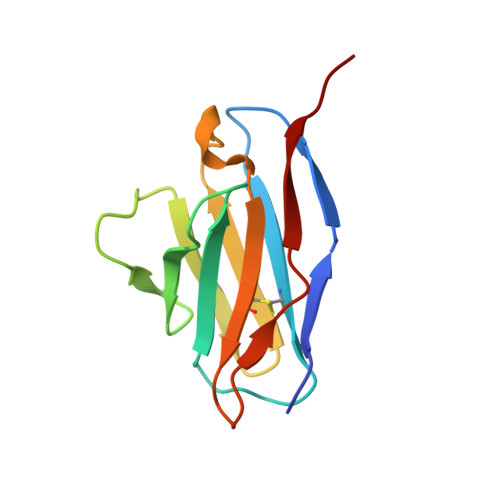
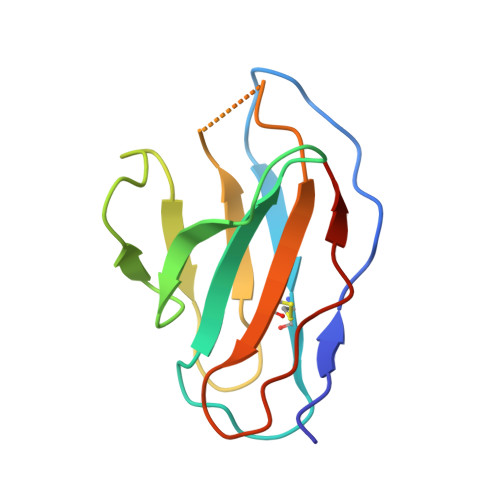
Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Ng, W.M., Fedosyuk, S., English, S., Augusto, G., Berg, A., Thorley, L., Haselon, A.S., Segireddy, R.R., Bowden, T.A., Douglas, A.D.(2022) Cell Host Microbe 30: 1219-1230.e7
- PubMed: 35985336
- DOI: https://doi.org/10.1016/j.chom.2022.07.014
- Primary Citation of Related Structures:
8A1E - PubMed Abstract:
Rabies virus (RABV) causes lethal encephalitis and is responsible for approximately 60,000 deaths per year. As the sole virion-surface protein, the rabies virus glycoprotein (RABV-G) mediates host-cell entry. RABV-G's pre-fusion trimeric conformation displays epitopes bound by protective neutralizing antibodies that can be induced by vaccination or passively administered for post-exposure prophylaxis. We report a 2.8-Å structure of a RABV-G trimer in the pre-fusion conformation, in complex with two neutralizing and protective monoclonal antibodies, 17C7 and 1112-1, that recognize distinct epitopes. One of these antibodies is a licensed prophylactic (17C7, Rabishield), which we show locks the protein in pre-fusion conformation. Targeted mutations can similarly stabilize RABV-G in the pre-fusion conformation, a key step toward structure-guided vaccine design. These data reveal the higher-order architecture of a key therapeutic target and the structural basis of neutralization by antibodies binding two key antigenic sites, and this will facilitate the development of improved vaccines and prophylactic antibodies.
- Jenner Institute, Old Road Campus Research Building, Roosevelt Drive, Oxford OX3 7DQ, UK; Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Organizational Affiliation: