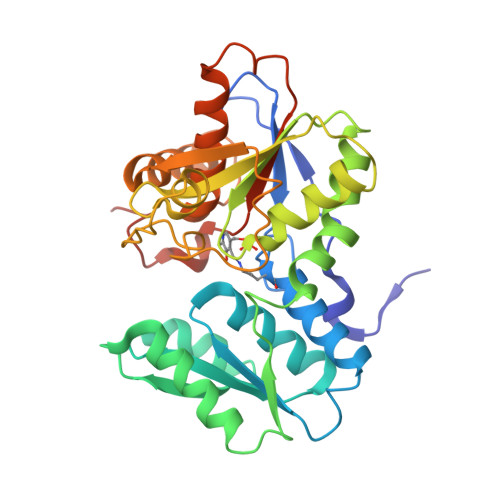
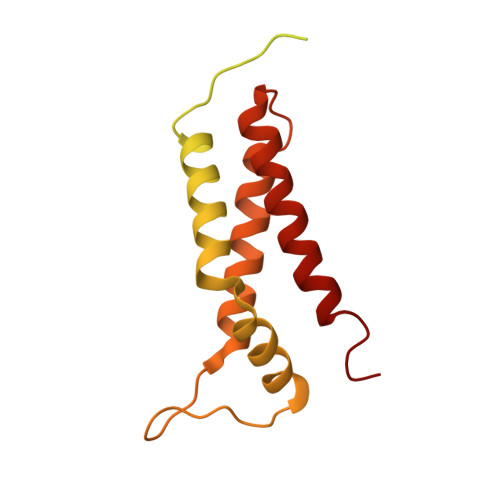
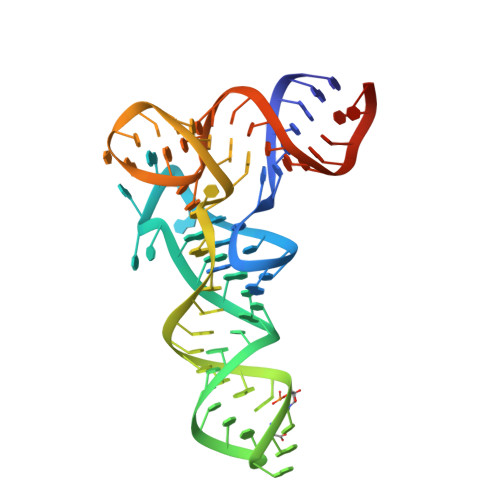
Mechanism of activation of contact-dependent growth inhibition tRNase toxin by the amino acid biogenesis factor CysK in the bacterial competition system.
Feng, Z., Yashiro, Y., Tomita, K.(2025) Nucleic Acids Res 53
- PubMed: 39228374
- DOI: https://doi.org/10.1093/nar/gkae735
- Primary Citation of Related Structures:
8ZYC, 8ZYD - PubMed Abstract:
Contact-dependent growth inhibition (CDI) is a bacterial competition mechanism, wherein the C-terminal toxin domain of CdiA protein (CdiA-CT) is transferred from one bacterium to another, impeding the growth of the toxin recipient. In uropathogenic Escherichia coli 536, CdiA-CT (CdiA-CTEC536) is a tRNA anticodon endonuclease that requires a cysteine biogenesis factor, CysK, for its activity. However, the mechanism underlying tRNA recognition and cleavage by CdiA-CTEC536 remains unresolved. Here, we present the cryo-EM structure of the CysK:CdiA-CTEC536:tRNA ternary complex. The interaction between CdiA-CTEC536 and CysK stabilizes the CdiA-CTEC536 structure and facilitates tRNA binding and the formation of the CdiA-CTEC536 catalytic core structure. The bottom-half of the tRNA interacts exclusively with CdiA-CTEC536 and the α-helices of CdiA-CTEC536 engage with the minor and major grooves of the bottom-half of tRNA, positioning the tRNA anticodon loop at the CdiA-CTEC536 catalytic site for tRNA cleavage. Thus, CysK serves as a platform facilitating the recognition and cleavage of substrate tRNAs by CdiA-CTEC536.
- Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.
Organizational Affiliation: