Structural basis of acetylcholine receptor like-23 (ACR-23) activation by anthelmintics monepantel and betaine
Liu, F.L., Li, T.Y., Gong, H.H., Guo, F., Liu, S., Chen, Q.F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Betaine receptor acr-23,Soluble cytochrome b562 | A [auth D], B [auth A], C [auth B], D [auth C], E | 620 | Caenorhabditis elegans, Escherichia coli | Mutation(s): 0 Gene Names: acr-23, 5E130, F59B1.9, cybC Membrane Entity: Yes | 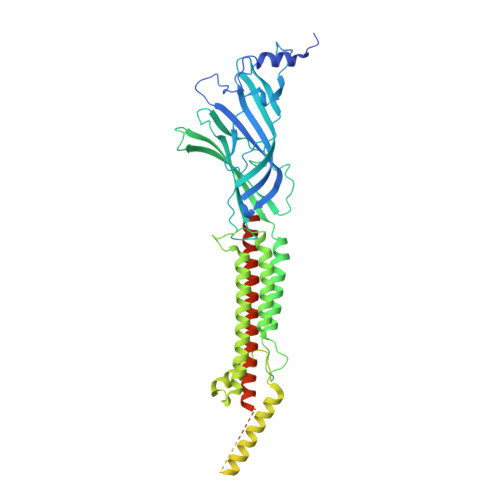 |
UniProt | |||||
Find proteins for G5EG88 (Caenorhabditis elegans) Explore G5EG88 Go to UniProtKB: G5EG88 | |||||
Find proteins for P0ABE7 (Escherichia coli) Explore P0ABE7 Go to UniProtKB: P0ABE7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P0ABE7G5EG88 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | P [auth D], R [auth A], T [auth B], V [auth C], X [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| BET (Subject of Investigation/LOI) Query on BET | Q [auth D], S [auth A], U [auth B], W [auth C], Y [auth E] | TRIMETHYL GLYCINE C5 H12 N O2 KWIUHFFTVRNATP-UHFFFAOYSA-O |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32071202 |
| National Natural Science Foundation of China (NSFC) | China | 32100129 |