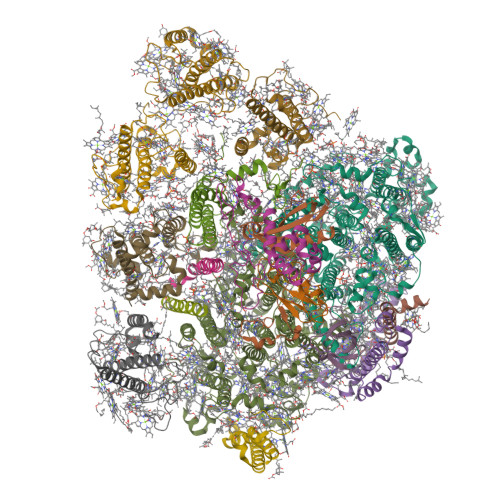
Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
Feng, Y., Li, Z., Yang, Y., Shen, L., Li, X., Liu, X., Zhang, X., Zhang, J., Ren, F., Wang, Y., Liu, C., Han, G., Wang, X., Kuang, T., Shen, J.R., Wang, W.(2024) J Integr Plant Biol
- PubMed: 39670505
- DOI: https://doi.org/10.1111/jipb.13816
- Primary Citation of Related Structures:
8ZEH, 8ZET - PubMed Abstract:
Diatoms rely on fucoxanthin chlorophyll a/c-binding proteins (FCPs) for light harvesting and energy quenching under marine environments. Here we report two cryo-electron microscopic structures of photosystem I (PSI) with either 13 or five fucoxanthin chlorophyll a/c-binding protein Is (FCPIs) at 2.78 and 3.20 Å resolutions from Thalassiosira pseudonana grown under high light (HL) conditions. Among them, five FCPIs are stably associated with the PSI core, these include Lhcr3, RedCAP, Lhcq8, Lhcf10, and FCP3. The eight additional Lhcr-type FCPIs are loosely associated with the PSI core and detached under the present purification conditions. The pigments of this centric diatom showed a higher proportion of chlorophylls a, diadinoxanthins, and diatoxanthins; some of the chlorophyll as and diadinoxanthins occupy the locations of fucoxanthins found in the huge PSI-FCPI from another centric diatom Chaetoceros gracilis grown under low-light conditions. These additional chlorophyll as may form more energy transfer pathways and additional diadinoxanthins may form more energy dissipation sites relying on the diadinoxanthin-diatoxanthin cycle. These results reveal the assembly mechanism of FCPIs and corresponding light-adaptive strategies of T. pseudonana PSI-FCPI, as well as the convergent evolution of the diatom PSI-FCPI structures.
Organizational Affiliation:
Key Laboratory of Photobiology, Photosynthesis Research Center, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China.