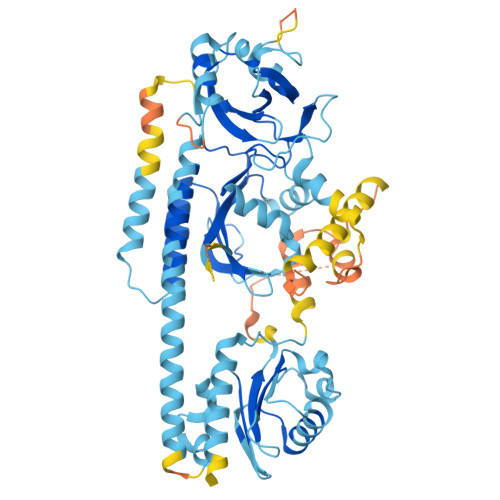
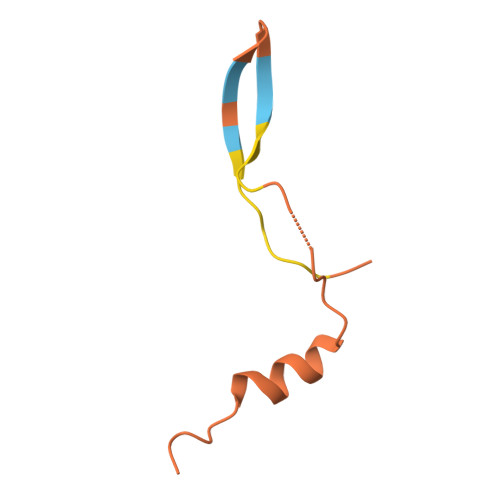
Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Wang, Z., Wang, W., Zhao, D., Song, Y., Lin, X., Shen, M., Chi, C., Xu, B., Zhao, J., Deng, X.W., Wang, J.(2024) Cell 187: 6235-6250.e19
- PubMed: 39317197
- DOI: https://doi.org/10.1016/j.cell.2024.09.005
- Primary Citation of Related Structures:
8YB4, 9IUZ - PubMed Abstract:
Phytochrome B (phyB) and phytochrome-interacting factors (PIFs) constitute a well-established signaling module critical for plants adapting to ambient light. However, mechanisms underlying phyB photoactivation and PIF binding for signal transduction remain elusive. Here, we report the cryo-electron microscopy (cryo-EM) structures of the photoactivated phyB or the constitutively active phyB Y276H mutant in complex with PIF6, revealing a similar trimer. The light-induced configuration switch of the chromophore drives a conformational transition of the nearby tongue signature within the phytochrome-specific (PHY) domain of phyB. The resulting α-helical PHY tongue further disrupts the head-to-tail dimer of phyB in the dark-adapted state. These structural remodelings of phyB facilitate the induced-fit recognition of PIF6, consequently stabilizing the N-terminal extension domain and a head-to-head dimer of activated phyB. Interestingly, the phyB dimer exhibits slight asymmetry, resulting in the binding of only one PIF6 molecule. Overall, our findings solve a key question with respect to how light-induced remodeling of phyB enables PIF signaling in phytochrome research.
Organizational Affiliation:
National Key Laboratory of Wheat Improvement, Peking University Institute of Advanced Agricultural Sciences, Shandong Laboratory of Advanced Agriculture Sciences at Weifang, Weifang, Shandong, China; State Key Laboratory of Protein and Plant Gene Research, School of Advanced Agricultural Sciences, Peking University, Beijing, China; Peking-Tsinghua Joint Center for Life Sciences, Peking University, Beijing, China.