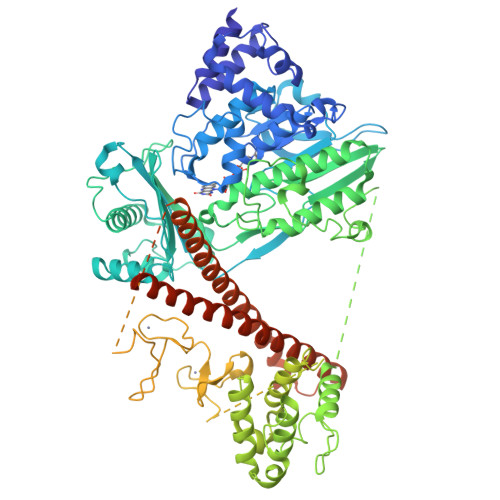
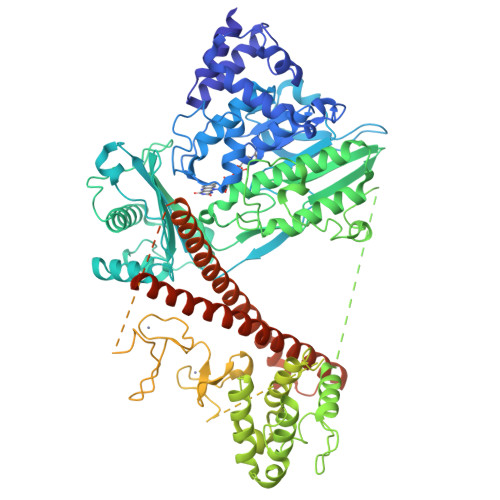
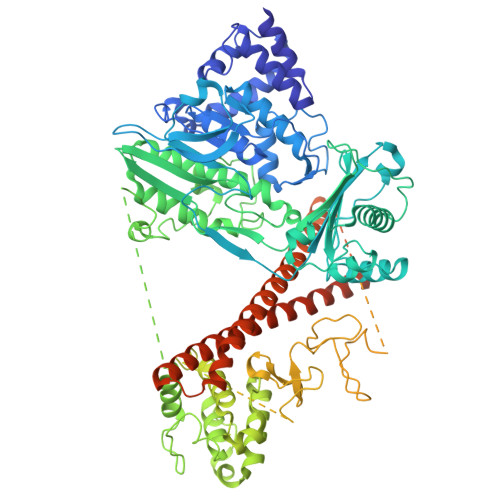
Autoinhibition and relief mechanisms for MICAL monooxygenases in F-actin disassembly.
Lin, L., Dong, J., Xu, S., Xiao, J., Yu, C., Niu, F., Wei, Z.(2024) Nat Commun 15: 6824-6824
- PubMed: 39122694
- DOI: https://doi.org/10.1038/s41467-024-50940-7
- Primary Citation of Related Structures:
8Y6K - PubMed Abstract:
MICAL proteins represent a unique family of actin regulators crucial for synapse development, membrane trafficking, and cytokinesis. Unlike classical actin regulators, MICALs catalyze the oxidation of specific residues within actin filaments to induce robust filament disassembly. The potent activity of MICALs requires tight control to prevent extensive damage to actin cytoskeleton. However, the molecular mechanism governing MICALs' activity regulation remains elusive. Here, we report the cryo-EM structure of MICAL1 in the autoinhibited state, unveiling a head-to-tail interaction that allosterically blocks enzymatic activity. The structure also reveals the assembly of C-terminal domains via a tripartite interdomain interaction, stabilizing the inhibitory conformation of the RBD. Our structural, biochemical, and cellular analyses elucidate a multi-step mechanism to relieve MICAL1 autoinhibition in response to the dual-binding of two Rab effectors, revealing its intricate activity regulation mechanisms. Furthermore, our mutagenesis study of MICAL3 suggests the conserved autoinhibition and relief mechanisms among MICALs.
Organizational Affiliation:
Shenzhen Key Laboratory of Biomolecular Assembling and Regulation, Shenzhen, Guangdong, China.