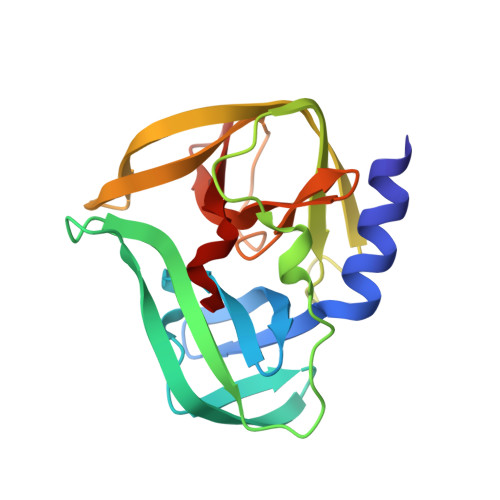
Crystal structures of the 3C proteases from Coxsackievirus B3 and B4.
Jiang, H., Lin, C., Chang, J., Zou, X., Zhang, J., Li, J.(2024) Acta Crystallogr F Struct Biol Commun 80: 183-190
- PubMed: 39052022
- DOI: https://doi.org/10.1107/S2053230X24006915
- Primary Citation of Related Structures:
8Y2T, 8Y2U - PubMed Abstract:
Enteroviruses cause a wide range of disorders with varying presentations and severities, and some enteroviruses have emerged as serious public health concerns. These include Coxsackievirus B3 (CVB3), an active causative agent of viral myocarditis, and Coxsackievirus B4 (CVB4), which may accelerate the progression of type 1 diabetes. The 3C proteases from CVB3 and CVB4 play important roles in the propagation of these viruses. In this study, the 3C proteases from CVB3 and CVB4 were expressed in Escherichia coli and purified by affinity chromatography and gel-filtration chromatography. The crystals of the CVB3 and CVB4 3C proteases diffracted to 2.10 and 2.01 Å resolution, respectively. The crystal structures were solved by the molecular-replacement method and contained a typical chymotrypsin-like fold and a conserved His40-Glu71-Cys147 catalytic triad. Comparison with the structures of 3C proteases from other enteroviruses revealed high similarity with minor differences, which will guide the design of 3C-targeting inhibitors with broad-spectrum properties.
Organizational Affiliation:
School of Basic Medical Sciences, Jiangxi Medical College, Nanchang University, Nanchang 330031, People's Republic of China.