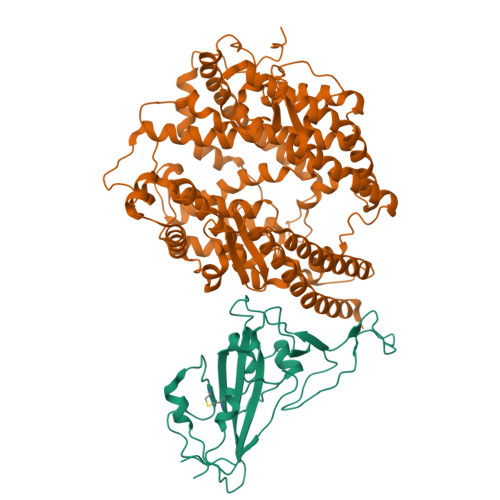
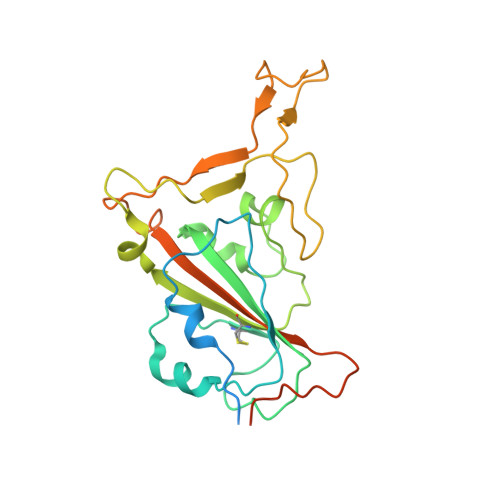
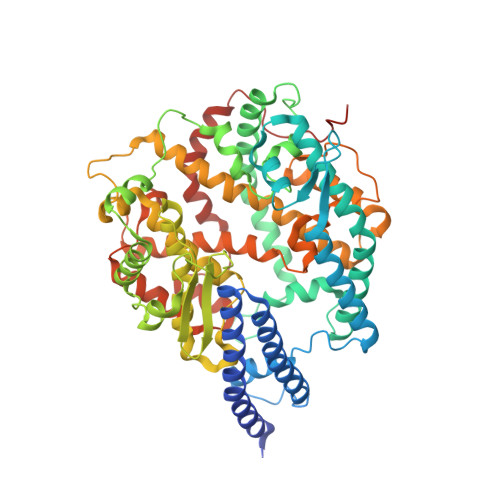
The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Chen, J., Sun, J., Xu, Z., Li, L., Kang, X., Luo, C., Wang, Q., Guo, X., Li, Y., Liu, K., Wu, Y.(2024) Virol Sin 39: 609-618
- PubMed: 38866203
- DOI: https://doi.org/10.1016/j.virs.2024.06.004
- Primary Citation of Related Structures:
8XYZ, 8XZB, 8XZD - PubMed Abstract:
Foxes are susceptible to SARS-CoV-2 in laboratory settings, and there have also been reports of natural infections of both SARS-CoV and SARS-CoV-2 in foxes. In this study, we assessed the binding capacities of fox ACE2 to important sarbecoviruses, including SARS-CoV, SARS-CoV-2, and animal-origin SARS-CoV-2 related viruses. Our findings demonstrated that fox ACE2 exhibits broad binding capabilities to receptor-binding domains (RBDs) of sarbecoviruses. We further determined the cryo-EM structures of fox ACE2 complexed with RBDs of SARS-CoV, SARS-CoV-2 prototype (PT), and Omicron BF.7. Through structural analysis, we identified that the K417 mutation can weaken the ability of SARS-CoV-2 sub-variants to bind to fox ACE2, thereby reducing the susceptibility of foxes to SARS-CoV-2 sub-variants. In addition, the Y498 residue in the SARS-CoV RBD plays a crucial role in forming a vital cation-π interaction with K353 in the fox ACE2 receptor. This interaction is the primary determinant for the higher affinity of the SARS-CoV RBD compared to that of the SARS-CoV-2 PT RBD. These results indicate that foxes serve as potential hosts for numerous sarbecoviruses, highlighting the critical importance of surveillance efforts.
Organizational Affiliation:
State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy and Immunology, Institute of Medical Virology, TaiKang Medical School (School of Basic Medical Sciences), Wuhan University, Wuhan, 430071, China.