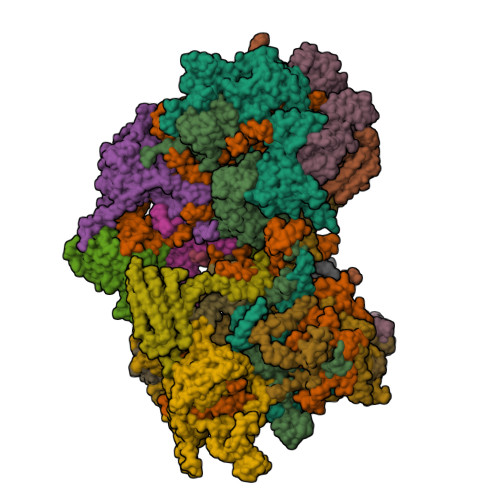
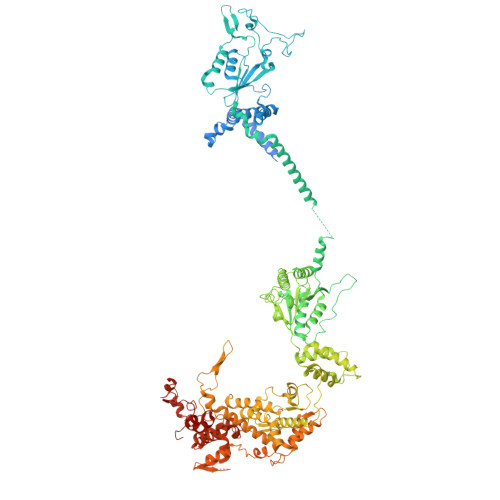
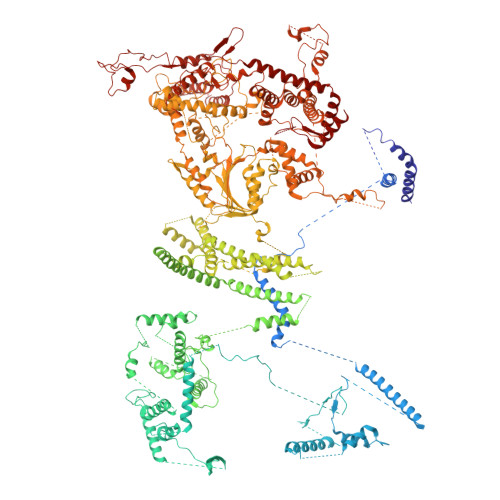
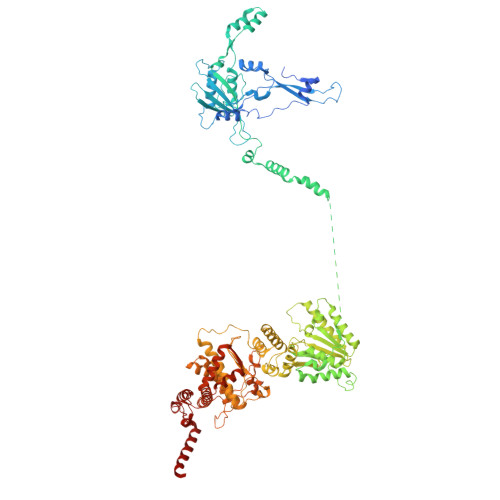
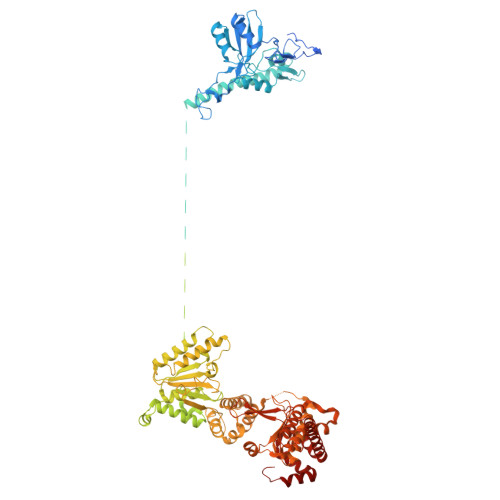
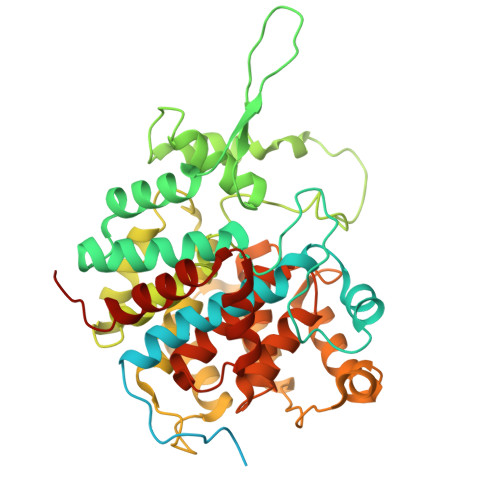
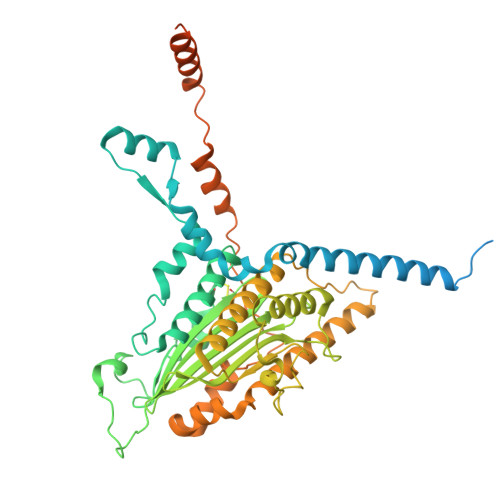
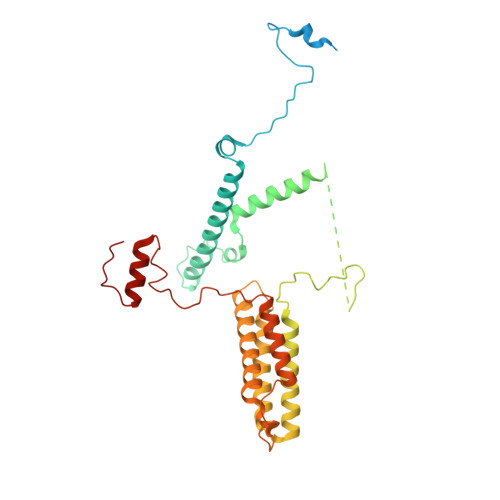
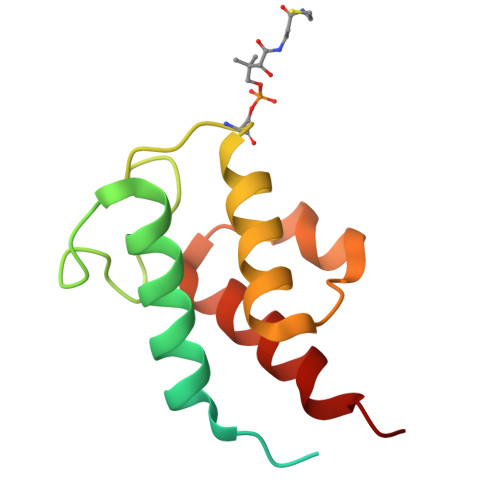
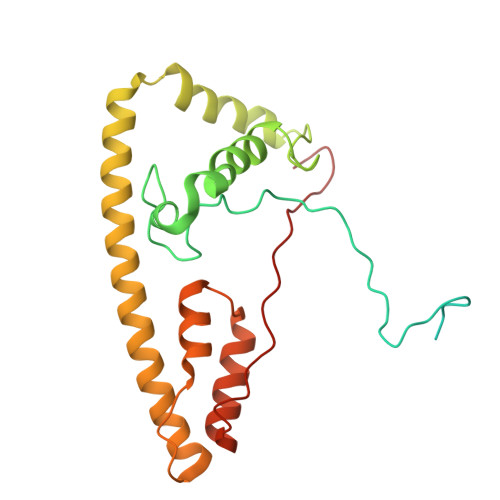
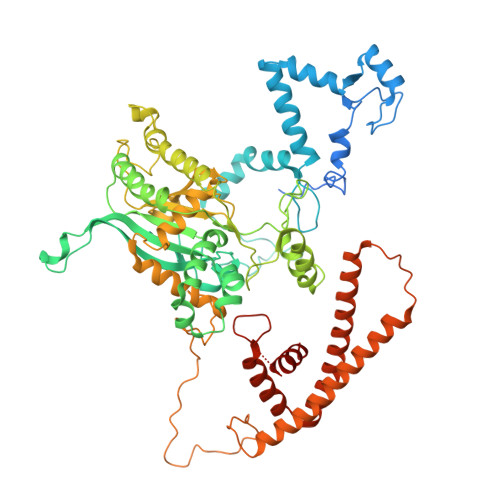
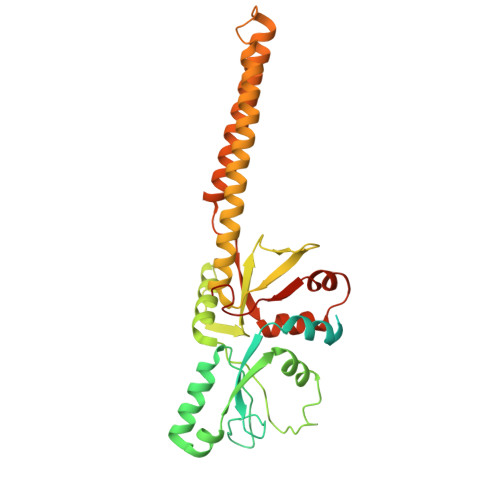
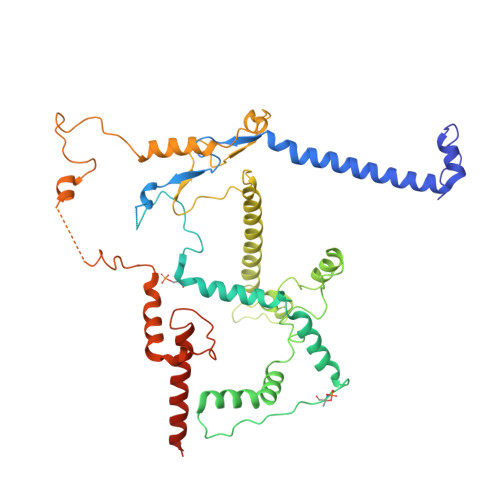
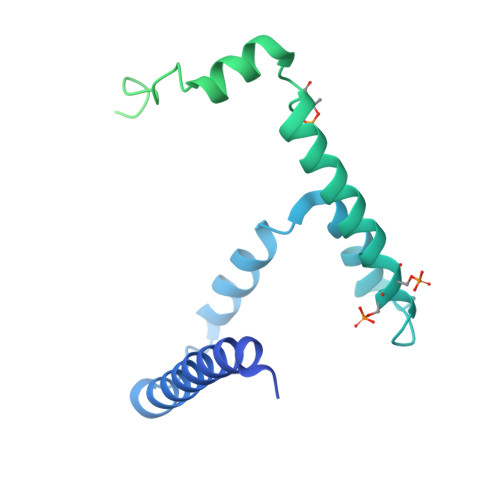
Conservation and specialization of the Ycf2-FtsHi chloroplast protein import motor in green algae.
Liang, K., Zhan, X., Li, Y., Yang, Y., Xie, Y., Jin, Z., Xu, X., Zhang, W., Lu, Y., Zhang, S., Zou, Y., Feng, S., Wu, J., Yan, Z.(2024) Cell 187: 5638-5650.e18
- PubMed: 39197449
- DOI: https://doi.org/10.1016/j.cell.2024.08.002
- Primary Citation of Related Structures:
8XQW, 8XQX - PubMed Abstract:
The protein import motor in chloroplasts plays a pivotal role in their biogenesis and homeostasis by driving the translocation of preproteins into chloroplasts. While the Ycf2-FtsHi complex serves as the import motor in land plants, its evolutionary conservation, specialization, and mechanisms across photosynthetic organisms are largely unexplored. Here, we isolated and determined the cryogenic electron microscopy (cryo-EM) structures of the native Ycf2-FtsHi complex from Chlamydomonas reinhardtii, uncovering a complex composed of up to 19 subunits, including multiple green-algae-specific components. The heterohexameric AAA+ ATPase motor module is tilted, potentially facilitating preprotein handover from the translocon at the inner chloroplast membrane (TIC) complex. Preprotein interacts with Ycf2-FtsHi and enhances its ATPase activity in vitro. Integrating Ycf2-FtsHi and translocon at the outer chloroplast membrane (TOC)-TIC supercomplex structures reveals insights into their physical and functional interplay during preprotein translocation. By comparing these findings with those from land plants, our study establishes a structural foundation for understanding the assembly, function, evolutionary conservation, and diversity of chloroplast protein import motors.
Organizational Affiliation:
College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang 310024, China; Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang 310024, China; Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, Zhejiang 310024, China; Institute of Biology, Westlake Institute for Advanced Study, Hangzhou, Zhejiang 310024, China.