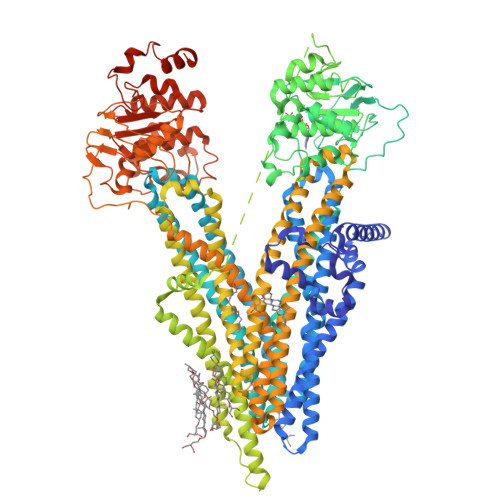
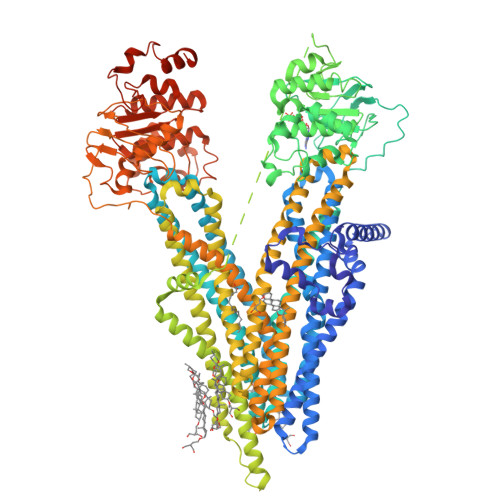
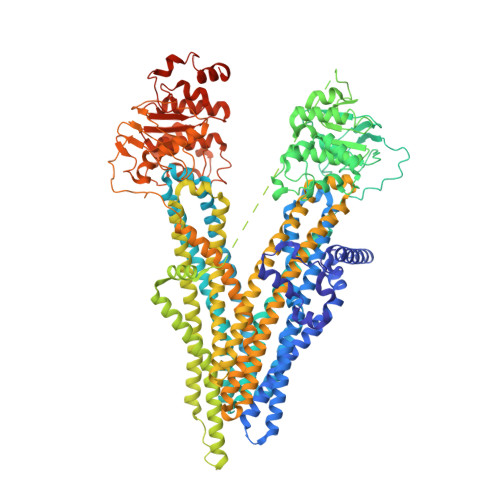
The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Zhu, Y., Xing, X., Wang, F., Chen, L., Zhong, C., Lu, X., Yu, Z., Yang, Y., Yao, Y., Song, Q., Han, S., Liu, Z., Zhang, P.(2024) FEBS Lett 598: 1967-1980
- PubMed: 38886124
- DOI: https://doi.org/10.1002/1873-3468.14955
- Primary Citation of Related Structures:
8XOK, 8XOL, 8XOM - PubMed Abstract:
The multidrug resistance-associated protein (MRP) ABCC4 facilitates substrate transport across the cytoplasmic membrane, crucial for normal physiology and mediating multidrug resistance in tumor cells. Despite intensive studies on MRPs, ABCC4's transport mechanism remains incompletely understood. In this study, we unveiled an inward-open conformation with an ATP bound to degenerate NBD1. Additionally, we captured the structure with both ATP and substrate co-bound in the inward-open state. Our findings uncover the asymmetric ATP binding in ABCC4 and provide insights into substrate binding and transport mechanisms. ATP binding to NBD1 is parallel to substrate binding to ABCC4, and is a prerequisite for ATP-bound NBD2-induced global conformational changes. Our findings shed new light on targeting ABCC4 in combination with anticancer therapy.
Organizational Affiliation:
Department of Radiation Oncology, The First Affiliated Hospital of Xi'an Jiaotong University, China.