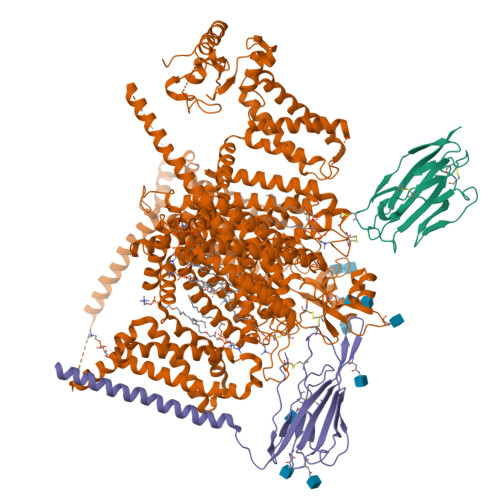
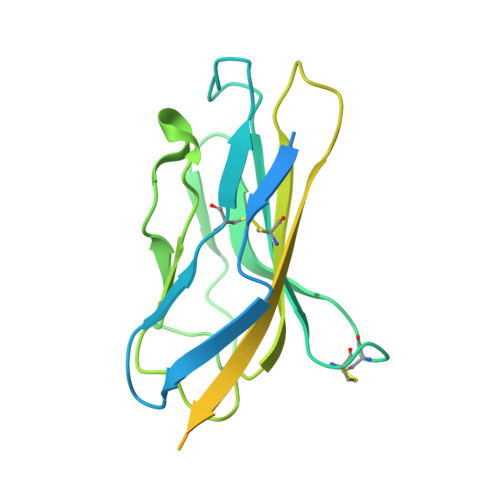
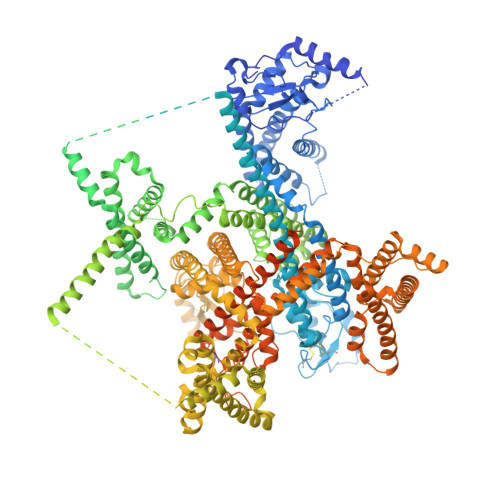
Dissection of the structure-function relationship of Na v channels.
Li, Z., Wu, Q., Huang, G., Jin, X., Li, J., Pan, X., Yan, N.(2024) Proc Natl Acad Sci U S A 121: e2322899121-e2322899121
- PubMed: 38381792
- DOI: https://doi.org/10.1073/pnas.2322899121
- Primary Citation of Related Structures:
8XMM, 8XMN, 8XMO - PubMed Abstract:
Voltage-gated sodium channels (Na v ) undergo conformational shifts in response to membrane potential changes, a mechanism known as the electromechanical coupling. To delineate the structure-function relationship of human Na v channels, we have performed systematic structural analysis using human Na v 1.7 as a prototype. Guided by the structural differences between wild-type (WT) Na v 1.7 and an eleven mutation-containing variant, designated Na v 1.7-M11, we generated three additional intermediate mutants and solved their structures at overall resolutions of 2.9-3.4 Å. The mutant with nine-point mutations in the pore domain (PD), named Na v 1.7-M9, has a reduced cavity volume and a sealed gate, with all voltage-sensing domains (VSDs) remaining up. Structural comparison of WT and Na v 1.7-M9 pinpoints two residues that may be critical to the tightening of the PD. However, the variant containing these two mutations, Na v 1.7-M2, or even in combination with two additional mutations in the VSDs, named Na v 1.7-M4, failed to tighten the PD. Our structural analysis reveals a tendency of PD contraction correlated with the right shift of the static inactivation I-V curves. We predict that the channel in the resting state should have a "tight" PD with down VSDs.
Organizational Affiliation:
Beijing Frontier Research Center for Biological Structures, State Key Laboratory of Membrane Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.