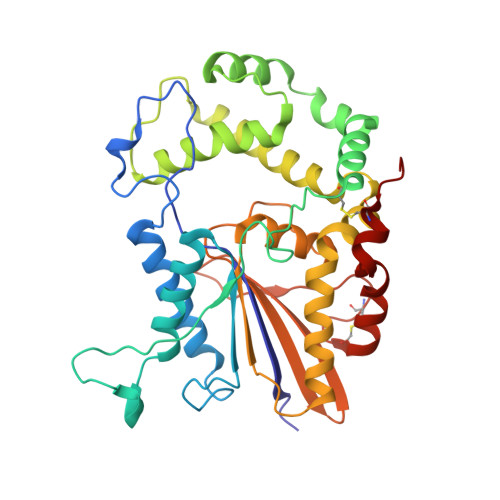
Purification, identification and Cryo-EM structure of prostatic acid phosphatase in human semen.
Liu, X., Yu, L., Xia, Z., Li, J., Meng, W., Min, L., Li, F., Wang, X.(2024) Biochem Biophys Res Commun 702: 149652-149652
- PubMed: 38341922
- DOI: https://doi.org/10.1016/j.bbrc.2024.149652
- Primary Citation of Related Structures:
8XJ4 - PubMed Abstract:
Prostatic acid phosphatase (PAP) is a glycoprotein that plays a crucial role in the hydrolysis of phosphate ester present in prostatic exudates. It is a well-established indicator for prostate cancer due to its elevated serum levels in disease progression. Despite its abundance in semen, PAP's influence on male fertility has not been extensively studied. In our study, we report a significantly optimized method for purifying human endogenous PAP, achieving remarkably high efficiency and active protein recovery rate. This achievement allowed us to better analyze and understand the PAP protein. We determined the cryo-electron microscopic (Cryo-EM) structure of prostatic acid phosphatase in its physiological state for the first time. Our structural and gel filtration analysis confirmed the formation of a tight homodimer structure of human PAP. This functional homodimer displayed an elongated conformation in the cryo-EM structure compared to the previously reported crystal structure. Additionally, there was a notable 5-degree rotation in the angle between the α domain and α/β domain of each monomer. Through structural analysis, we revealed three potential glycosylation sites: Asn94, Asn220, and Asn333. These sites contained varying numbers and forms of glycosyl units, suggesting sugar moieties influence PAP function. Furthermore, we found that the active sites of PAP, His44 and Asp290, are located between the two protein domains. Overall, our study not only provide an optimized approach for PAP purification, but also offer crucial insights into its structural characteristics. These findings lay the groundwork for further investigations into the physiological function and potential therapeutic applications of this important protein.
Organizational Affiliation:
Department of Obstetrics and Gynecology, Key Laboratory of Birth Defects and Related Disease of Women and Children of MOE, State Key Laboratory of Biotherapy, West China Second Hospital, Sichuan University, Chengdu, 610041, Sichuan Province, China.