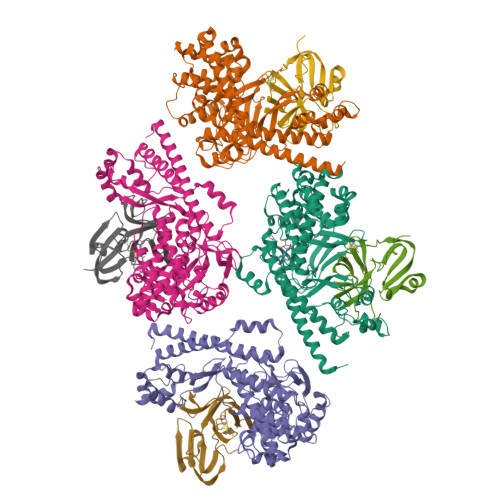
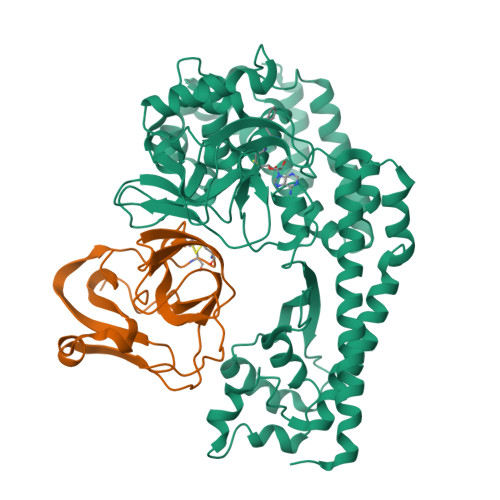
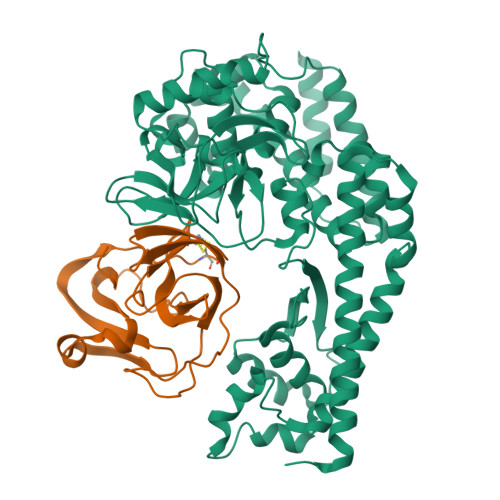
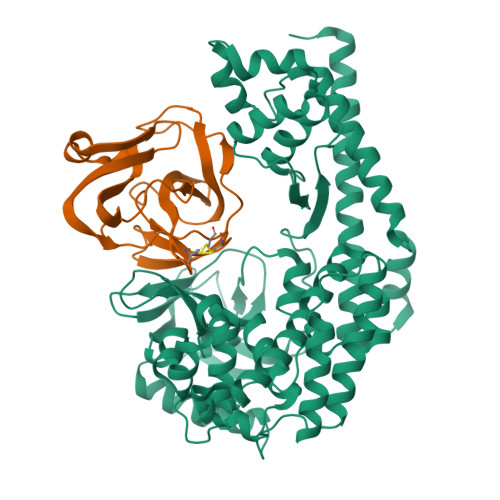
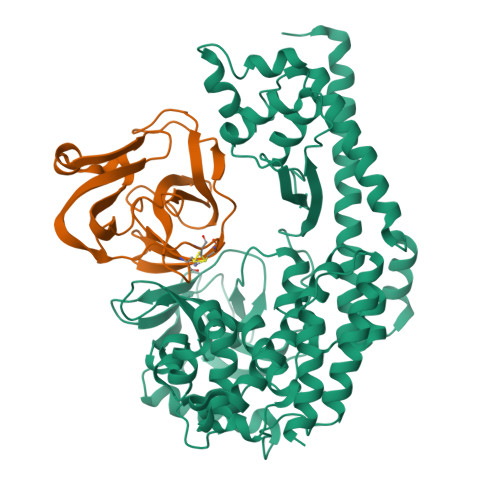
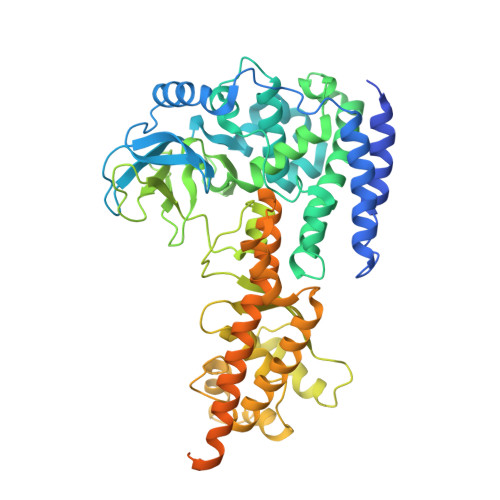
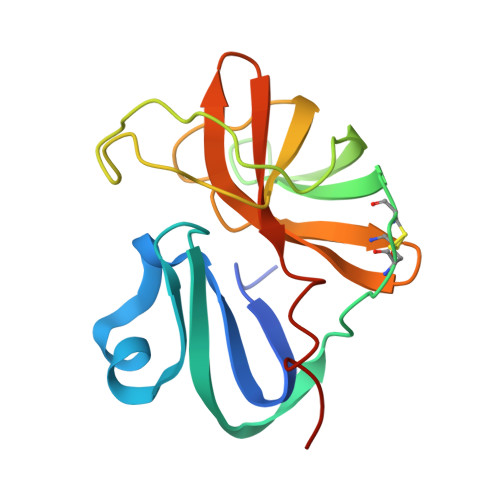
The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Gao, X., Wang, B., Zhu, K., Wang, L., Qin, B., Shang, K., Ding, W., Wang, J., Cui, S.(2024) Nat Commun 15: 4176-4176
- PubMed: 38755176
- DOI: https://doi.org/10.1038/s41467-024-48504-w
- Primary Citation of Related Structures:
8X77, 8X8Q - PubMed Abstract:
SETD3 is an essential host factor for the replication of a variety of enteroviruses that specifically interacts with viral protease 2A. However, the interaction between SETD3 and the 2A protease has not been fully characterized. Here, we use X-ray crystallography and cryo-electron microscopy to determine the structures of SETD3 complexed with the 2A protease of EV71 to 3.5 Å and 3.1 Å resolution, respectively. We find that the 2A protease occupies the V-shaped central cleft of SETD3 through two discrete sites. The relative positions of the two proteins vary in the crystal and cryo-EM structures, showing dynamic binding. A biolayer interferometry assay shows that the EV71 2A protease outcompetes actin for SETD3 binding. We identify key 2A residues involved in SETD3 binding and demonstrate that 2A's ability to bind SETD3 correlates with EV71 production in cells. Coimmunoprecipitation experiments in EV71 infected and 2A expressing cells indicate that 2A interferes with the SETD3-actin complex, and the disruption of this complex reduces enterovirus replication. Together, these results reveal the molecular mechanism underlying the interplay between SETD3, actin, and viral 2A during virus replication.
Organizational Affiliation:
NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100730, China.