Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Yuan, L., Liu, Q., Xu, L., Wu, B., Feng, Y.(2024) Nat Commun 15: 4850-4850
- PubMed: 38844782
- DOI: https://doi.org/10.1038/s41467-024-49229-6
- Primary Citation of Related Structures:
8X6F, 8X6G - PubMed Abstract:
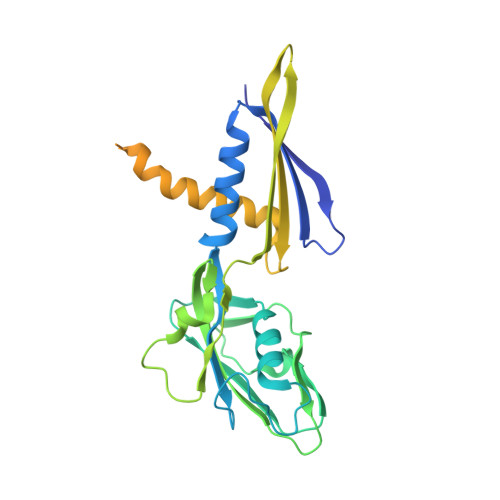
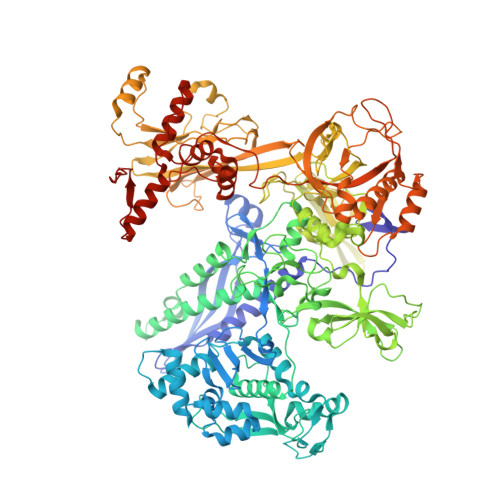
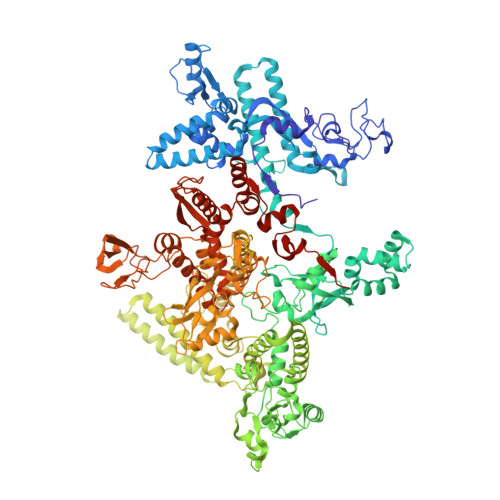
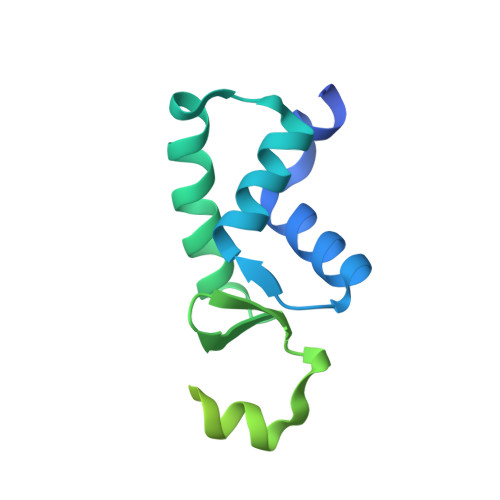
Bacterial RNAP needs to form holoenzyme with σ factors to initiate transcription. While Staphylococcus aureus σ A controls housekeeping functions, S. aureus σ B regulates virulence, biofilm formation, persistence, cell internalization, membrane transport, and antimicrobial resistance. Besides the sequence difference, the spacers between the -35 element and -10 element of σ B regulated promoters are shorter than those of σ A regulated promoters. Therefore, how σ B recognizes and initiates transcription from target promoters can not be inferred from that of the well studied σ. Here, we report the cryo-EM structures of S. aureus RNAP-promoter open complexes comprising σ A and σ B , respectively. Structural analyses, in combination with biochemical experiments, reveal the structural basis for the promoter specificity of S. aureus transcription. Although the -10 element of σ A regulated promoters is recognized by domain σ A 2 as single-stranded DNA, the -10 element of σ B regulated promoters is co-recognized by domains σ B 2 and σ B 3 as double-stranded DNA, accounting for the short spacers of σ B regulated promoters. S. aureus RNAP is a validated target of antibiotics, and our structures pave the way for rational drug design targeting S. aureus RNAP.
Organizational Affiliation:
Department of Biophysics, and Department of Infectious Disease of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China.