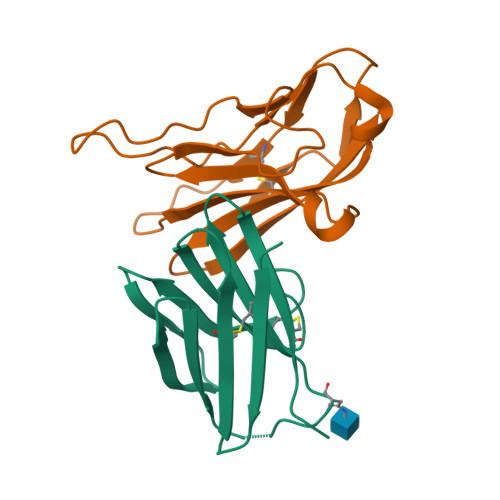
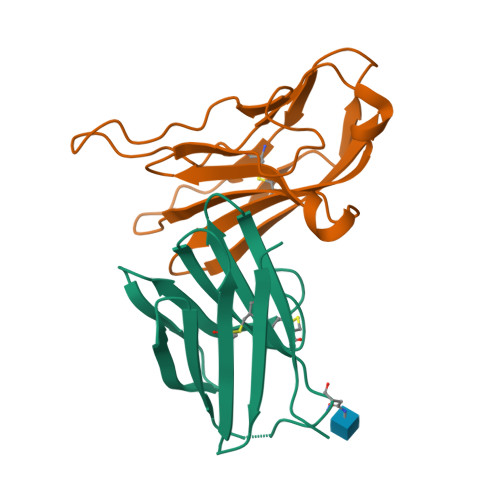
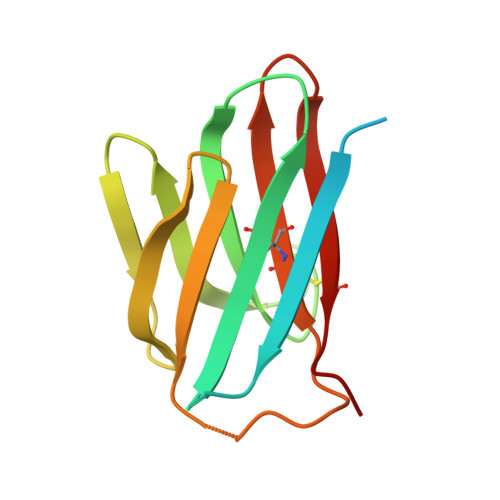
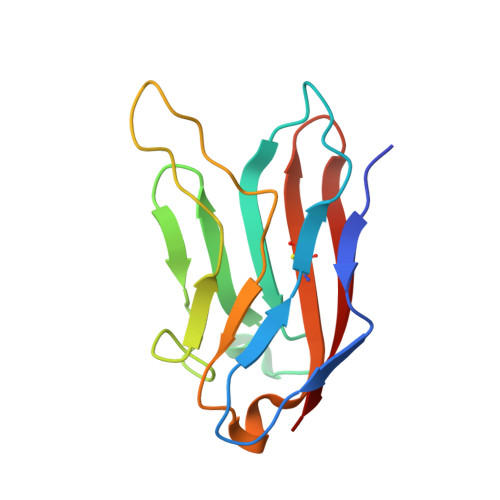
Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Hu, S., Han, P., Wang, M., Cao, X., Liu, H., Zhang, S., Zhang, S., Liu, J., Han, Y., Xiao, J., Chen, Q., Miao, K., Qi, J., Tan, S., Gao, G.F., Wang, H.(2024) Structure 32: 918-929.e4
- PubMed: 38626767
- DOI: https://doi.org/10.1016/j.str.2024.03.012
- Primary Citation of Related Structures:
8X6B - PubMed Abstract:
Nectin and nectin-like (Necl) co-receptor axis, comprised of receptors DNAM-1, TIGIT, CD96, PVRIG, and nectin/Necl ligands, is gaining prominence in immuno-oncology. Within this axis, the inhibitory receptor PVRIG recognizes Nectin-2 with high affinity, but the underlying molecular basis remains unknown. By determining the crystal structure of PVRIG in complex with Nectin-2, we identified a unique CC' loop in PVRIG, which complements the double-lock-and-key binding mode and contributes to its high affinity for Nectin-2. The association of the corresponding charged residues in the F-strands explains the ligand selectivity of PVRIG toward Nectin-2 but not for Necl-5. Moreover, comprehensive comparisons of the binding capacities between co-receptors and ligands provide innovative insights into the intra-axis immunoregulatory mechanism. Taken together, these findings broaden our understanding of immune recognition and regulation mediated by nectin/Necl co-receptors and provide a rationale for the development of immunotherapeutic strategies targeting the nectin/Necl axis.
Organizational Affiliation:
Institutes of Physical Science and Information Technology, Anhui University, Anhui 230601, China; Cancer Center, Faculty of Health Sciences, University of Macau, Taipa Macau SAR, China; Beijing Life Science Academy, Beijing 102200, China.