Overall structure of the LAT1-4F2hc bound with L-dopa
Yan, R.H., Li, Y.N., Shi, T.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Amino acid transporter heavy chain SLC3A2 | 470 | Homo sapiens | Mutation(s): 0 Gene Names: SLC3A2, MDU1 Membrane Entity: Yes | 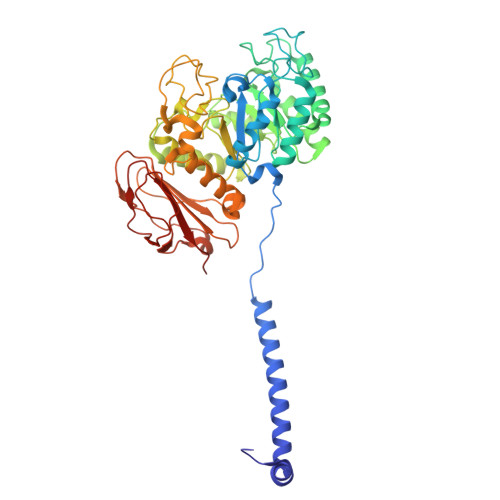 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08195 (Homo sapiens) Explore P08195 Go to UniProtKB: P08195 | |||||
PHAROS: P08195 GTEx: ENSG00000168003 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08195 | ||||
Glycosylation | |||||
| Glycosylation Sites: 4 | Go to GlyGen: P08195-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Large neutral amino acids transporter small subunit 1 | 464 | Homo sapiens | Mutation(s): 0 Gene Names: SLC7A5, CD98LC, LAT1, MPE16 Membrane Entity: Yes | 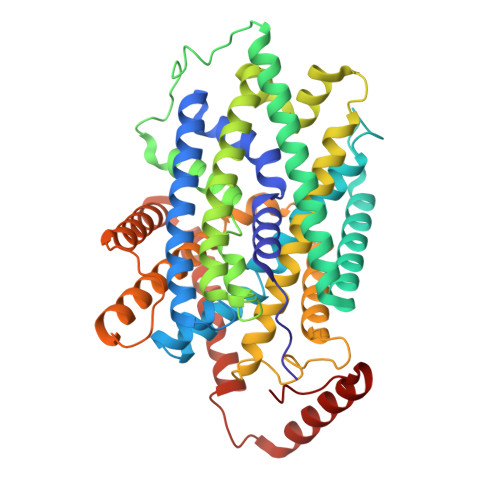 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q01650 (Homo sapiens) Explore Q01650 Go to UniProtKB: Q01650 | |||||
PHAROS: Q01650 GTEx: ENSG00000103257 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q01650 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LEU Query on LEU | G [auth B] | LEUCINE C6 H13 N O2 ROHFNLRQFUQHCH-YFKPBYRVSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |