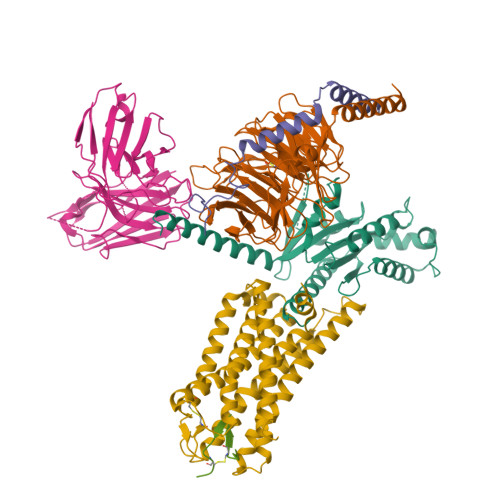
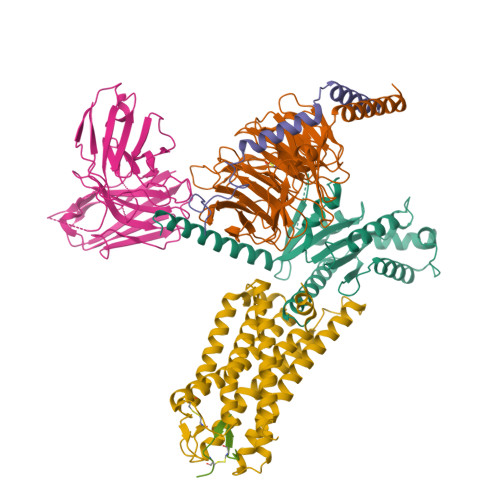
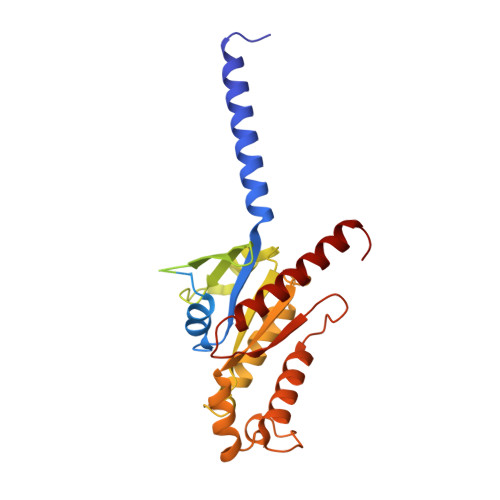
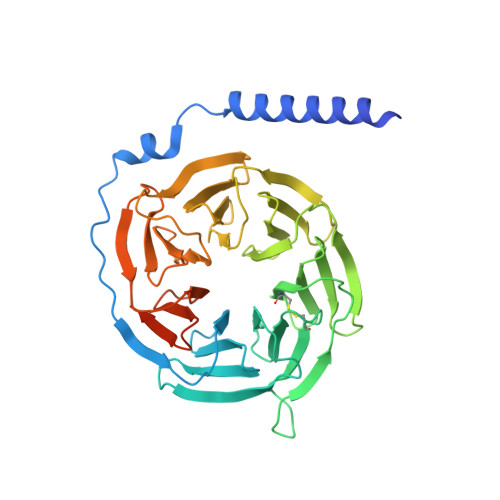
Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
He, Q., Yuan, Q., Shan, H., Wu, C., Gu, Y., Wu, K., Hu, W., Zhang, Y., He, X., Xu, H.E., Zhao, L.H.(2024) Cell Discov 10: 48-48
- PubMed: 38710677
- DOI: https://doi.org/10.1038/s41421-024-00679-8
- Primary Citation of Related Structures:
8WSS, 8WST - PubMed Abstract:
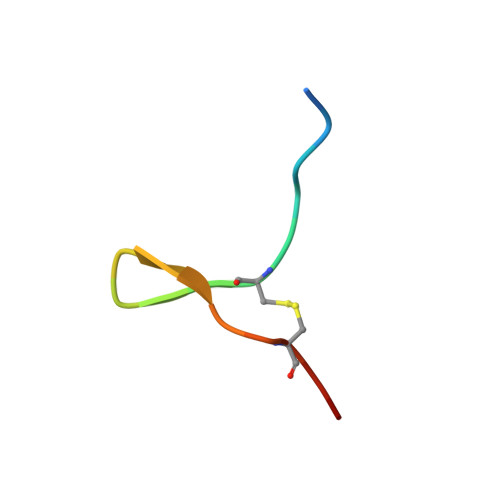
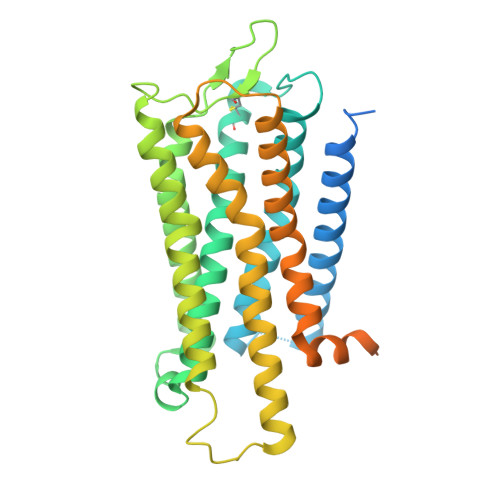
Melanin-concentrating hormone (MCH) is a cyclic neuropeptide that regulates food intake, energy balance, and other physiological functions by stimulating MCHR1 and MCHR2 receptors, both of which are class A G protein-coupled receptors. MCHR1 predominately couples to inhibitory G protein, G i/o , and MCHR2 can only couple to G q/11 . Here we present cryo-electron microscopy structures of MCH-activated MCHR1 with G i and MCH-activated MCHR2 with G q at the global resolutions of 3.01 Å and 2.40 Å, respectively. These structures reveal that MCH adopts a consistent cysteine-mediated hairpin loop configuration when bound to both receptors. A central arginine from the LGRVY core motif between the two cysteines of MCH penetrates deeply into the transmembrane pocket, triggering receptor activation. Integrated with mutational and functional insights, our findings elucidate the molecular underpinnings of ligand recognition and MCH receptor activation and offer a structural foundation for targeted drug design.
Organizational Affiliation:
State Key Laboratory of Drug Research, Center for Structure and Function of Drug Targets, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.