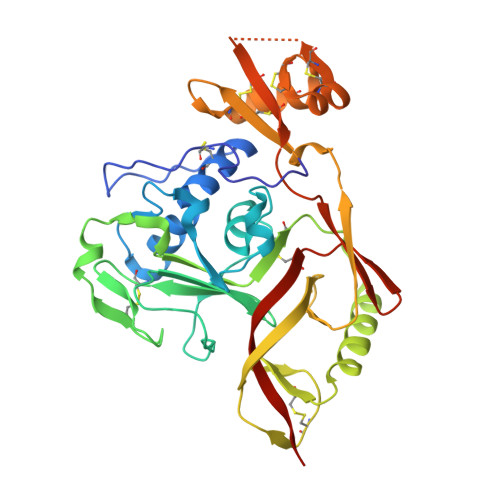
Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Chang, Z., Gao, D., Liao, L., Sun, J., Zhang, G., Zhang, X., Wang, F., Li, C., Oladejo, B.O., Li, S., Chai, Y., Hu, Y., Lu, X., Xiao, H., Qi, J., Chen, Z., Gao, F., Wu, Y.(2024) Proc Natl Acad Sci U S A 121: e2400163121-e2400163121
- PubMed: 38830098
- DOI: https://doi.org/10.1073/pnas.2400163121
- Primary Citation of Related Structures:
8WQW, 8WSN, 8WSP, 8WSU - PubMed Abstract:
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease with a high fatality rate of up to 30% caused by SFTS virus (SFTSV). However, no specific vaccine or antiviral therapy has been approved for clinical use. To develop an effective treatment, we isolated a panel of human monoclonal antibodies (mAbs). SF5 and SF83 are two neutralizing mAbs that recognize two viral glycoproteins (Gn and Gc), respectively. We found that their epitopes are closely located, and we then engineered them as several bispecific antibodies (bsAbs). Neutralization and animal experiments indicated that bsAbs display more potent protective effects than the parental mAbs, and the cryoelectron microscopy structure of a bsAb3 Fab-Gn-Gc complex elucidated the mechanism of protection. In vivo virus passage in the presence of antibodies indicated that two bsAbs resulted in less selective pressure and could efficiently bind to all single parental mAb-escape mutants. Furthermore, epitope analysis of the protective mAbs against SFTSV and RVFV indicated that they are all located on the Gn subdomain I, where may be the hot spots in the phleboviruses. Collectively, these data provide potential therapeutic agents and molecular basis for the rational design of vaccines against SFTSV infection.
Organizational Affiliation:
Department of Pathogen Microbiology, School of Basic Medical Sciences, Capital Medical University, Beijing 100069, China.