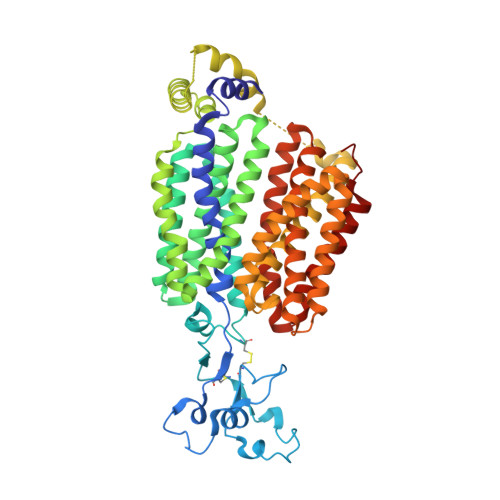
Structural basis for the transport and substrate selection of human urate transporter 1.
He, J., Liu, G., Kong, F., Tan, Q., Wang, Z., Yang, M., He, Y., Jia, X., Yan, C., Wang, C., Qian, H.(2024) Cell Rep 43: 114628-114628
- PubMed: 39146184
- DOI: https://doi.org/10.1016/j.celrep.2024.114628
- Primary Citation of Related Structures:
8WJG, 8WJH, 8WJQ - PubMed Abstract:
High serum urate levels are the major risk factor for gout. URAT1, the primary transporter for urate absorption in the kidneys, is well known as an anti-hyperuricemia drug target. However, the clinical application of URAT1-targeted drugs is limited because of their low specificity and severe side effects. The lack of structural information impedes elucidation of the transport mechanism and the development of new drugs. Here, we present the cryoelectron microscopy (cryo-EM) structures of human URAT1(R477S), its complex with urate, and its closely related homolog OAT4. URAT1(R477S) and OAT4 exhibit major facilitator superfamily (MFS) folds with outward- and inward-open conformations, respectively. Structural comparison reveals a 30° rotation between the N-terminal and C-terminal domains, supporting an alternating access mechanism. A conserved arginine (OAT4-Arg473/URAT1-Arg477) is found to be essential for chloride-mediated inhibition. The URAT1(R477S)-urate complex reveals the specificity of urate recognition. Taken together, our study promotes our understanding of the transport mechanism and substrate selection of URAT1.
Organizational Affiliation:
Department of Cardiology, First Affiliated Hospital of USTC, MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Research Center for Interdisciplinary Sciences at the Microscale, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China.