Degradation of Polo-like Kinase 1 by the Novel Poly-Arginine N-Degron Pathway PROTAC Regulates Tumor Growth in Nonsmall Cell Lung Cancer.
Gunasekaran, P., Hwang, Y.S., Lee, G.H., Park, J., Kim, J.G., La, Y.K., Park, N.Y., Kothandaraman, R., Yim, M.S., Choi, J., Kim, H.N., Park, I.Y., Lee, S.J., Kim, M.H., Cha-Molstad, H., Shin, S.Y., Ryu, E.K., Bang, J.K.(2024) J Med Chem 67: 3307-3320
- PubMed: 38105611
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01493
- Primary Citation of Related Structures:
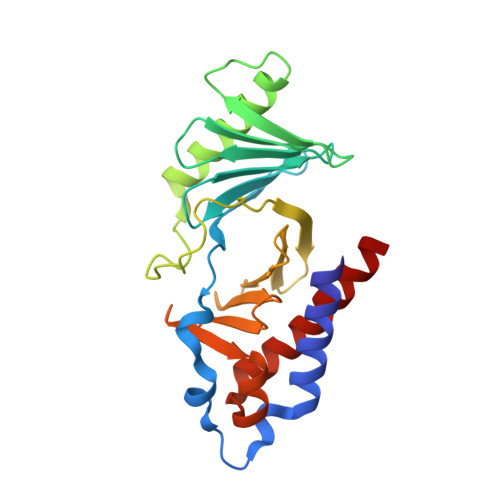
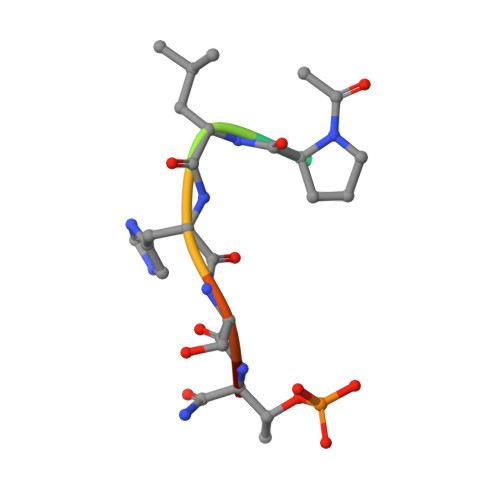
8WFP - PubMed Abstract:
Polo-like kinase 1 (PLK1), which is crucial in cell cycle regulation, is considered a promising anticancer drug target. Herein, we present the N-degron pathway-based proteolysis targeting chimera (PROTAC) for PLK1 degradation, targeting the Polo-box domain (PBD). We identified DD-2 as the most potent PROTAC that selectively induces PLK1 degradation in cancer cells, including HeLa and nonsmall cell lung cancer (NSCLC), through the N-degron pathway. DD-2 exhibited significant in vitro anticancer effects, inducing G2/M arrest and apoptosis in HeLa and NSCLC cell lines. DD-2 showed significant tumor growth inhibition in a xenograft mouse model using HeLa and NSCLC cell lines, highlighting its potential in cancer treatment. Furthermore, the combination of DD-2 with tyrosine kinase inhibitor (TKI), osimertinib, effectively suppressed tumor growth in double-mutated H1975 cell lines, emphasizing DD-2's potential in combination cancer therapies. Collectively, this study demonstrates the potential of the N-degron pathway, especially using DD-2, for targeted cancer therapies.
Organizational Affiliation:
Division of Magnetic Resonance, Korea Basic Science Institute (KBSI), Ochang, Chungbuk 28119, Republic of Korea.