Structural Characterization of an N-Acetyl Sugar Amidotransferase Involved in the Lipopolysaccharide Biosynthesis in Bacteria.
Gao, J., Xu, W., Liu, T., Sun, W., Wang, N., Ma, J., Ge, H.(2023) Int J Mol Sci 24
- PubMed: 37895170
- DOI: https://doi.org/10.3390/ijms242015491
- Primary Citation of Related Structures:
8WEX - PubMed Abstract:
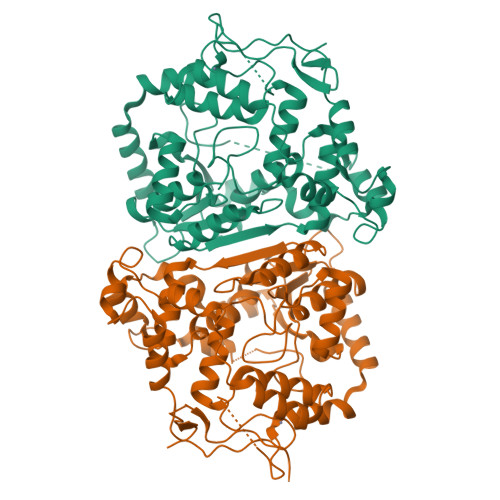
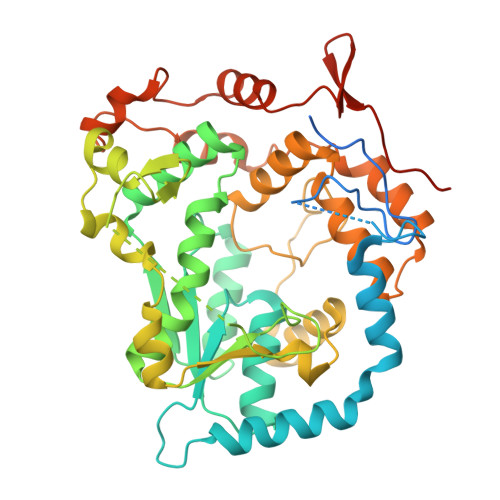
N-acetyl sugar amidotransferase (NASAT) is involved in the lipopolysaccharide (LPS) biosynthesis pathway that catalyzes the formation of the acetamido moiety (sugar-NC(=NH)CH3) on the O-chain. So far, little is known about its structural and functional properties. Here, we report the crystal structure of an N-acetyl sugar amidotransferase from Legionella pneumophila (LpNASAT) at 2.33 Å resolution. LpNASAT folds into a compact basin-shaped architecture with an unusually wide and open putative substrate-binding pocket and a conserved zinc ion-binding tetracysteine motif. The pocket contains a Rossmann-like fold with a PP-loop, suggesting that the NASAT-catalyzed amidotransfer reaction probably requires the conversion of ATP to AMP and PPi. Our data provide structural insights into the NASAT family of proteins, and allow us to possibly identify its functionally important regions.
Organizational Affiliation:
Information Materials and Intelligent Sensing Laboratory of Anhui Province, Institutes of Material Science and Information Technology, Anhui University, Hefei 230601, China.