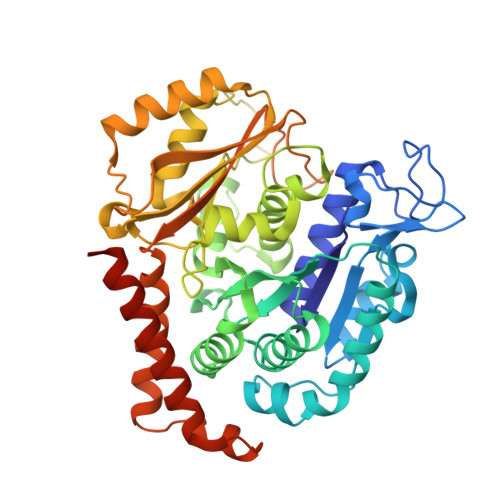
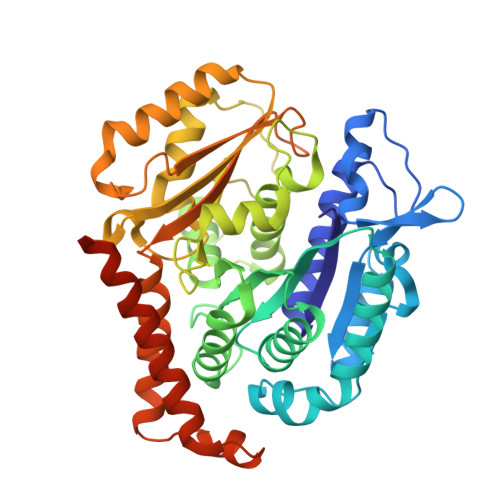
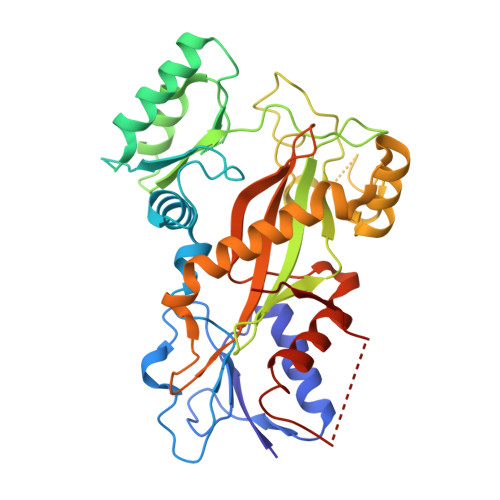
The cytotoxic natural compound erianin binds to colchicine site of beta-tubulin and overcomes taxane resistance
Yan, W., Zhou, Y., Yuan, X., Bai, P., Tang, M., Chen, L., Wei, H., Yang, J.(2024) Bioorg Chem 150: 107569
- PubMed: 38905886
- DOI: https://doi.org/10.1016/j.bioorg.2024.107569
- Primary Citation of Related Structures:
8WD0 - PubMed Abstract:
Erianin, a natural compound derived from Dendrobium, has shown significant anticancer properties against a wide range of cancer cells. Despite the identification of multiple mechanisms of action for erianin, none of these mechanisms fully account for its broad-spectrum effect. In this study, we aimed to identify the cellular target and underlying mechanism responsible for the broad-spectrum antitumor effects of erianin. We found that erianin effectively inhibited tubulin polymerization in cancer cells and purified tubulin. Through competition binding assays and X-ray crystallography, it was revealed that erianin bound to the colchicine site of β-tubulin. Importantly, the X-ray crystal structure of the tubulin-erianin complex was solved, providing clear insight into the orientation and position of erianin in the colchicine-binding site. Erianin showed activity against paclitaxel-resistant cells, evidenced by G2/M cell cycle arrest, apoptosis-related PARP and Caspase-3 cleavage, and in vivo xenograft studies. The study concluded that erianin bound reversibly to the colchicine site of β-tubulin, inhibited tubulin polymerization, and displayed anticancer activity against paclitaxel-resistant cells, offering valuable insights for further exploration as potential anticancer agents.
Organizational Affiliation:
Department of Biotherapy, Cancer Center and State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu 610041, China. Electronic address: yanxiaowei369@sina.com.