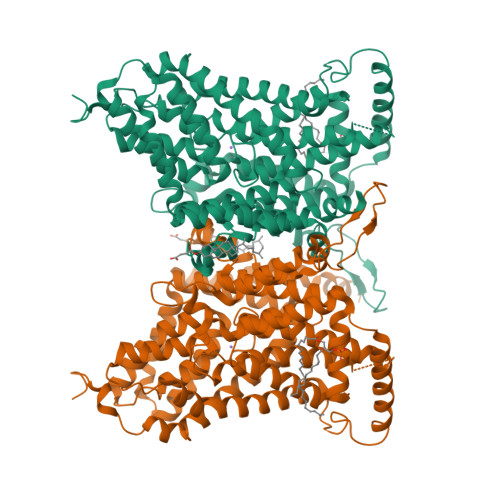
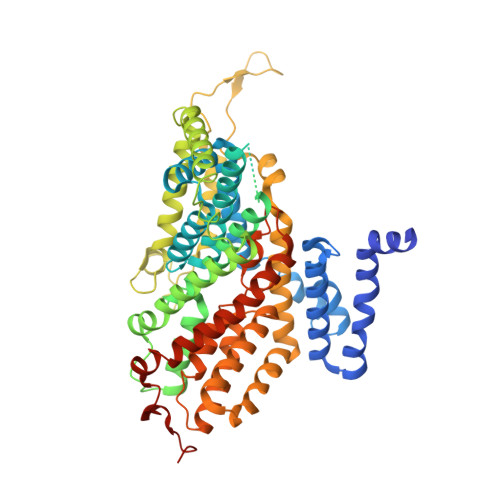
Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Chi, X., Chen, Y., Li, Y., Dai, L., Zhang, Y., Shen, Y., Chen, Y., Shi, T., Yang, H., Wang, Z., Yan, R.(2024) Sci Adv 10: eadl3685-eadl3685
- PubMed: 38552027
- DOI: https://doi.org/10.1126/sciadv.adl3685
- Primary Citation of Related Structures:
8W6C, 8W6D, 8W6G, 8W6H, 8W6N, 8W6O, 8W6T - PubMed Abstract:
The solute carrier 13 (SLC13) family comprises electrogenic sodium ion-coupled anion cotransporters, segregating into sodium ion-sulfate cotransporters (NaSs) and sodium ion-di- and-tricarboxylate cotransporters (NaDCs). NaS1 and NaDC1 regulate sulfate homeostasis and oxidative metabolism, respectively. NaS1 deficiency affects murine growth and fertility, while NaDC1 affects urinary citrate and calcium nephrolithiasis. Despite their importance, the mechanisms of substrate recognition and transport remain insufficiently characterized. In this study, we determined the cryo-electron microscopy structures of human NaS1, capturing inward-facing and combined inward-facing/outward-facing conformations within a dimer both in apo and sulfate-bound states. In addition, we elucidated NaDC1's outward-facing conformation, encompassing apo, citrate-bound, and N -( p -amylcinnamoyl) anthranilic acid (ACA) inhibitor-bound states. Structural scrutiny illuminates a detailed elevator mechanism driving conformational changes. Notably, the ACA inhibitor unexpectedly binds primarily anchored by transmembrane 2 (TM2), Loop 10, TM11, and TM6a proximate to the cytosolic membrane. Our findings provide crucial insights into SLC13 transport mechanisms, paving the way for future drug design.
Organizational Affiliation:
State Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Signaling Network, School of Life Science, Xiamen University, Xiamen 361102, Fujian Province, China.