Human Serum is a Potent Chemoattractant for Enterobacteriaceae Species Associated with Gastrointestinal Bleeding
Glenn, S., Gentry-Lear, Z., Shavlik, M., Harms, M.J., Asaki, T.J., Baylink, A.(2024) Elife
Experimental Data Snapshot
(2024) Elife
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methyl-accepting chemotaxis protein | 163 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: G0L35_02695 | 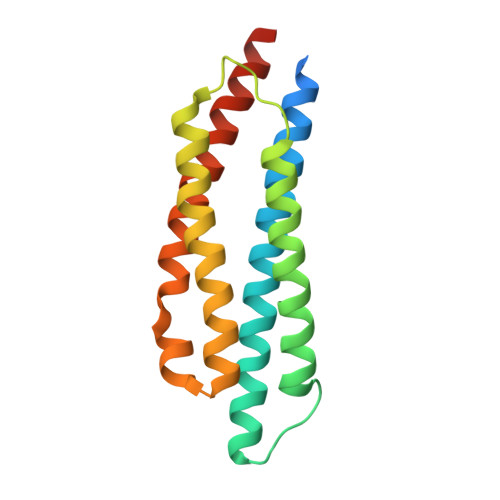 | |
UniProt | |||||
Find proteins for Q8ZJY4 (Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)) Explore Q8ZJY4 Go to UniProtKB: Q8ZJY4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8ZJY4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SER (Subject of Investigation/LOI) Query on SER | BA [auth E], G [auth A], R [auth B], V [auth C], Z [auth D] | SERINE C3 H7 N O3 MTCFGRXMJLQNBG-REOHCLBHSA-N |  | ||
| K Query on K | J [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| CL Query on CL | H [auth A] I [auth A] P [auth B] Q [auth B] U [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | AA [auth E] F [auth A] K [auth B] L [auth B] M [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 126.022 | α = 90 |
| b = 74.771 | β = 116.14 |
| c = 128.931 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | 5R00AI148587 |