Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction
Guercio, A.M., Gilio, A.K., Pawlak, J., Shabek, N.(2024) J Biol Chem
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2024) J Biol Chem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable esterase D14L | 272 | Oryza sativa | Mutation(s): 0 Gene Names: D14L, Os03g0437600, LOC_Os03g32270, Os11g0384789 | 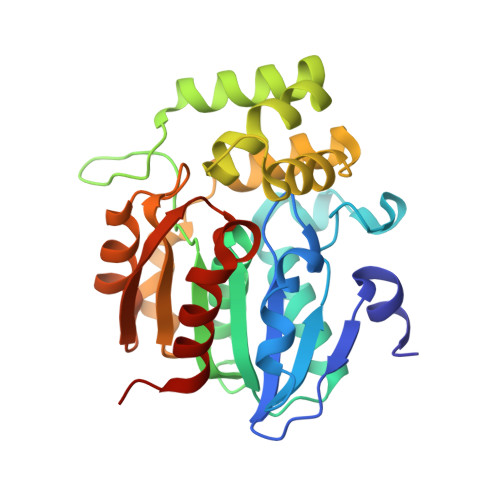 | |
UniProt | |||||
Find proteins for Q10J20 (Oryza sativa subsp. japonica) Explore Q10J20 Go to UniProtKB: Q10J20 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q10J20 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 37.636 | α = 90 |
| b = 85.385 | β = 114.91 |
| c = 44.62 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | -- |