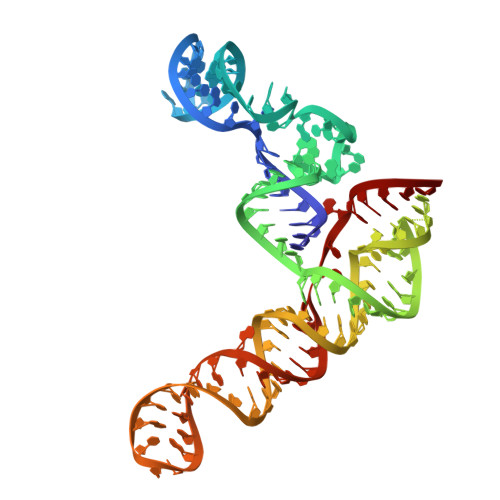
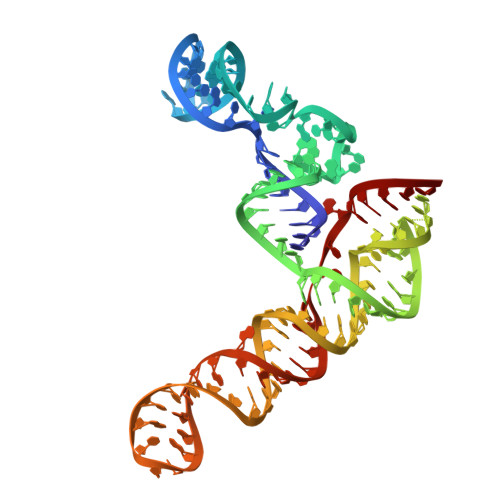
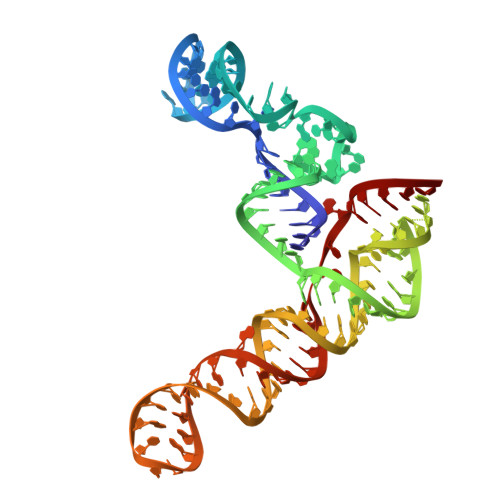
Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Peterson, J.M., Becker, S.T., O'Leary, C.A., Juneja, P., Yang, Y., Moss, W.N.(2024) Biochemistry 63: 1287-1296
- PubMed: 38727003
- DOI: https://doi.org/10.1021/acs.biochem.3c00716
- Primary Citation of Related Structures:
8VCI - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshift stimulatory element (FSE) is necessary for programmed -1 ribosomal frameshifting (-1 PRF) and optimized viral efficacy. The FSE has an abundance of context-dependent alternate conformations, but two of the structures most crucial to -1 PRF are an attenuator hairpin and a three-stem H-type pseudoknot structure. A crystal structure of the pseudoknot alone features three RNA stems in a helically stacked linear structure, whereas a 6.9 Å cryo-EM structure including the upstream heptameric slippery site resulted in a bend between two stems. Our previous research alluded to an extended upstream multibranch loop that includes both the attenuator hairpin and the slippery site-a conformation not previously modeled. We aim to provide further context to the SARS-CoV-2 FSE via computational and medium resolution cryo-EM approaches, by presenting a 6.1 Å cryo-EM structure featuring a linear pseudoknot structure and a dynamic upstream multibranch loop.
Organizational Affiliation:
Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, Iowa 50011, United States.