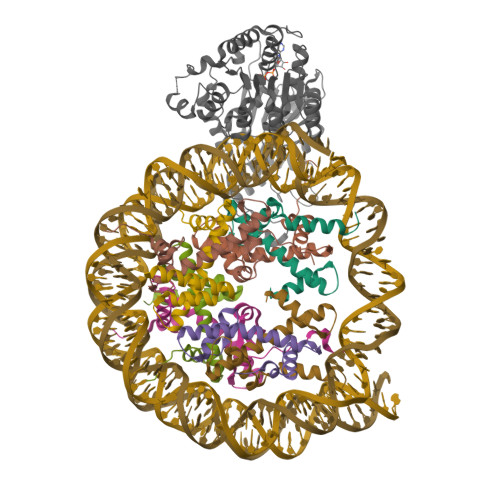
Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Chio, U.S., Palovcak, E., Smith, A.A.A., Autzen, H., Munoz, E.N., Yu, Z., Wang, F., Agard, D.A., Armache, J.P., Narlikar, G.J., Cheng, Y.(2024) Nat Commun 15: 2225-2225
- PubMed: 38472177
- DOI: https://doi.org/10.1038/s41467-024-46178-y
- Primary Citation of Related Structures:
8V4Y, 8V6V, 8V7L - PubMed Abstract:
Single-particle cryo-EM is widely used to determine enzyme-nucleosome complex structures. However, cryo-EM sample preparation remains challenging and inconsistent due to complex denaturation at the air-water interface (AWI). Here, to address this issue, we develop graphene-oxide-coated EM grids functionalized with either single-stranded DNA (ssDNA) or thiol-poly(acrylic acid-co-styrene) (TAASTY) co-polymer. These grids protect complexes between the chromatin remodeler SNF2h and nucleosomes from the AWI and facilitate collection of high-quality micrographs of intact SNF2h-nucleosome complexes in the absence of crosslinking. The data yields maps ranging from 2.3 to 3 Å in resolution. 3D variability analysis reveals nucleotide-state linked conformational changes in SNF2h bound to a nucleosome. In addition, the analysis provides structural evidence for asymmetric coordination between two SNF2h protomers acting on the same nucleosome. We envision these grids will enable similar detailed structural analyses for other enzyme-nucleosome complexes and possibly other protein-nucleic acid complexes in general.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, CA, USA.