Architecture and activation mechanism of the bacterial PARIS defence system
Deep, A., Liang, Q., Enustun, E., Pogliano, J., Corbett, K.D.(2024) Nature
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Nature
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AriA antitoxin | A, B, D [auth C], E [auth F] | 464 | Escherichia coli B185 | Mutation(s): 1 Gene Names: ECDG_03487 | 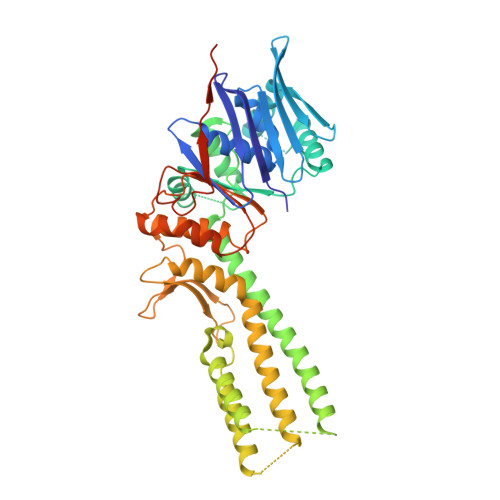 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AriB | C [auth D] | 308 | Escherichia coli B185 | Mutation(s): 1 Gene Names: ECDG_03486 | 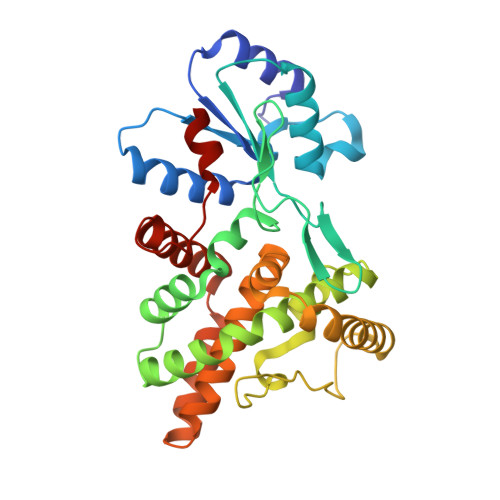 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | F [auth A], H [auth B] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | G [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21rc1_5127 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM144121 |