Structural Insights into Capsid Conformation and Dynamics: Cryo-EM Structures of HBV Capsids from HEK-293T Cells
Bianchini, E.N., Liu, H., Cai, Y., Shanklin, J., Hu, J., Wang, J.C.Y.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Capsid protein | 183 | Hepatitis B virus | Mutation(s): 3 Gene Names: pre-c/core, C, core, pre-C/C, PreC, preC/C, precore-core, HBVgp4 | 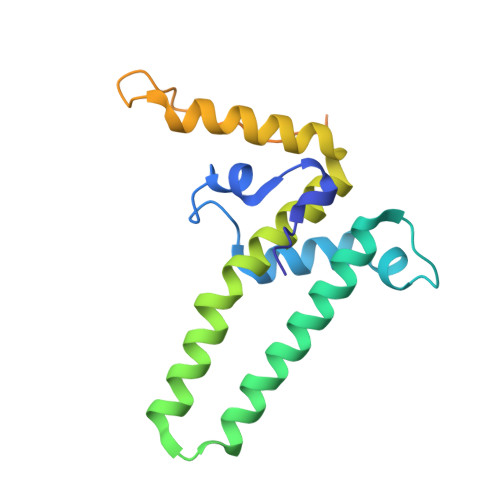 | |
UniProt | |||||
Find proteins for Q89656 (Hepatitis B virus) Explore Q89656 Go to UniProtKB: Q89656 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q89656 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | ISOLDE | |
| MODEL REFINEMENT | PHENIX | |
| MODEL REFINEMENT | Coot | |
| RECONSTRUCTION | RELION | 3.1 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI173104 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | AI043453 |