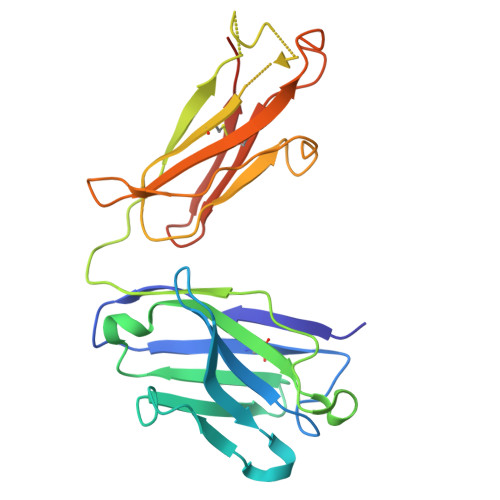
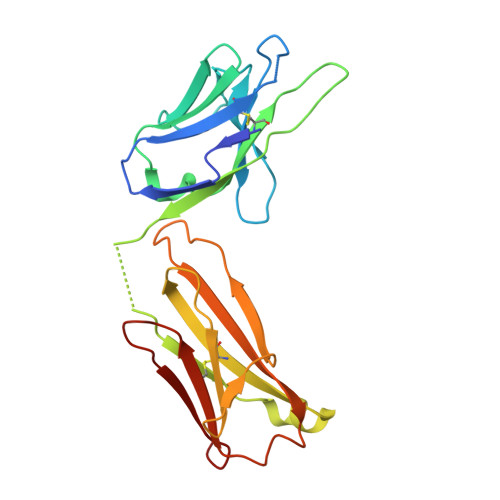
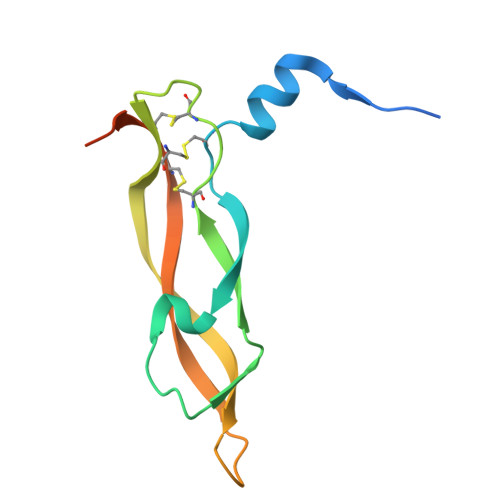
Raamsizumab: A Potent Synthetic Human Anti-VEGF Neutralizing Fab for Treatment of Ocular Neovascularization Diseases
Fellouse, F.A., Harit, R.K., Arun, S., Arora, R., Wagner, N., Antony, A., Wong, S., Yang, N., Rajakrishnan, S., Anthony, M., Gokul, S., Vinodh, S., Bhunia, S., Vinothini, T.M., Choubey, S., Rajendran, A., Srivastava, A., Seshagiri, S., Gross, M.L., Leung, D.W., Amarasinghe, G.K., Chaudhuri, A., Sidhu, S.S., Ramchand, C.N.To be published.