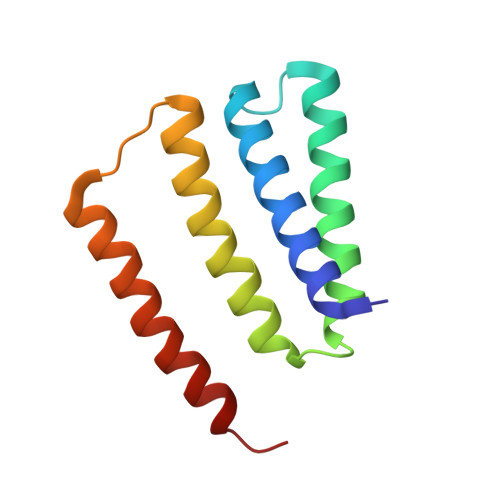
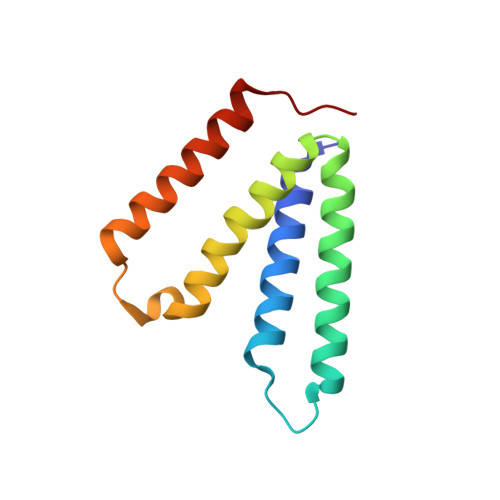
Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Li, J., Her, A.S., Besch, A., Ramirez-Cordero, B., Crames, M., Banigan, J.R., Mueller, C., Marsiglia, W.M., Zhang, Y., Traaseth, N.J.(2024) Nat Commun 15: 4537-4537
- PubMed: 38806470
- DOI: https://doi.org/10.1038/s41467-024-48803-2
- Primary Citation of Related Structures:
8UOZ, 8UWU - PubMed Abstract:
The multidrug efflux transporter EmrE from Escherichia coli requires anionic residues in the substrate binding pocket for coupling drug transport with the proton motive force. Here, we show how protonation of a single membrane embedded glutamate residue (Glu14) within the homodimer of EmrE modulates the structure and dynamics in an allosteric manner using NMR spectroscopy. The structure of EmrE in the Glu14 protonated state displays a partially occluded conformation that is inaccessible for drug binding by the presence of aromatic residues in the binding pocket. Deprotonation of a single Glu14 residue in one monomer induces an equilibrium shift toward the open state by altering its side chain position and that of a nearby tryptophan residue. This structural change promotes an open conformation that facilitates drug binding through a conformational selection mechanism and increases the binding affinity by approximately 2000-fold. The prevalence of proton-coupled exchange in efflux systems suggests a mechanism that may be shared in other antiporters where acid/base chemistry modulates access of drugs to the substrate binding pocket.
Organizational Affiliation:
Department of Chemistry, New York University, New York, NY, USA.